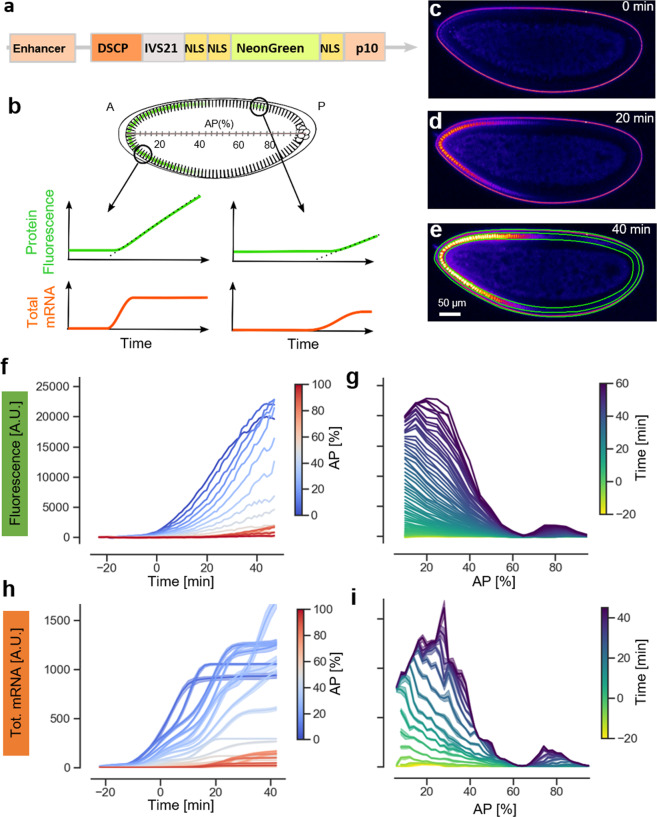
Fig. 1. A mNeonGreen reporter system to measure enhancer expression in living Drosophila melanogaster embryos.
a Composition of the mNeonGreen reporter construct including: Enhancer, basal DSCP promoter, the translation enhancer sequence IVS21 at the 5′UTR, the mNeonGreen fused to three nuclear localization signals and the terminator sequence p10. b Illustration of the use of a fluorescent protein as a transcriptional reporter. The time course of protein fluorescence (in green) is different in different portions of the embryo and carries information on the underlying dynamics of the total mRNA production (in orange), which can be computed using the reconstruction algorithm described in Supplementary Fig. 1. c–e Representative confocal slices of embryos carrying hb_ant-mNeonRep at three different time points during embryo development, showing mNeonGreen fluorescence in false colors. f Fluorescence time course traces corresponding to the average signal in 2% bins along the AP axis of the embryo, color coded by their position along the axis. g Fluorescence expression patterns along the AP axis. Each track corresponds to a different time of embryo development. h Time course of cumulative mRNA production in 2% bins along the AP axis of the embryo, color coded by their position along the axis. i Total mRNA production patterns at selected times of embryo development.