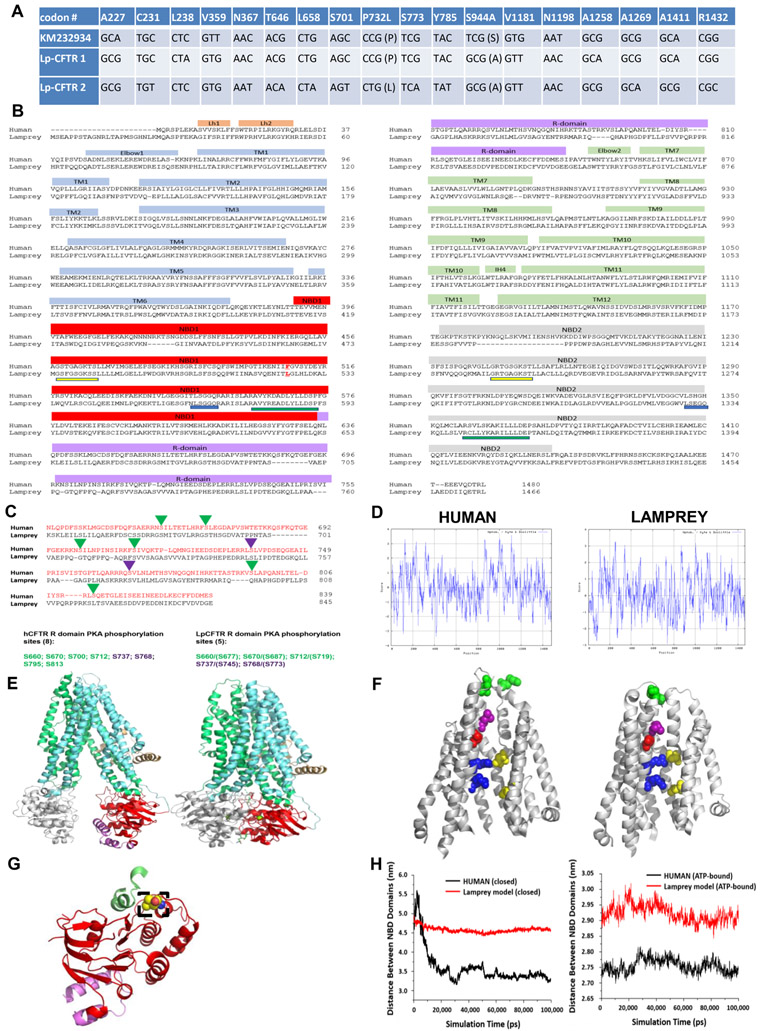
Figure 1: Lamprey CFTR (Lp-CFTR) demonstrates key similarities and differences in sequence and structure compared to human CFTR (hCFTR).
A: SNPs identified in Lp-CFTR. The Lp-CFTR gene was cloned from lamprey cDNA, sequenced, and compared to a published putative sequence (KM232934). Eighteen SNPs were identified with 2 amino acid changes between the three Lp-CFTR sequences.
B: Amino acid sequence of Lp-CFTR aligned with that of hCFTR. Key major domains are delineated above alignment. Lp-CFTR has an extended N-terminus, a leucine at the position corresponding to F508 in hCFTR (delineated by red letter and underline), intact Walker A domains (yellow underline) and Walker B domains (green underline) in both NBD1 and NBD2, but conserved signature sequence (blue lines) in NBD1 only.
C: Lp-CFTR has a reduced number of consensus PKA phosphorylation sites. The R-domains of Lp-CFTR and hCFTR were aligned and predicted phosphorylation sites were identified for activation (green arrowheads) and inhibition (purple arrowheads), reflecting a reduced number of consensus PKA phosphorylation sites in Lp-CFTR.
D: Similar hydropathy predicted from lamprey and human CFTR sequences. CFTR sequences were compared using the Kyte-Doolittle scale for hydropathy via ExPasy, with increased hydrophilicity as negative values and increased hydrophobicity as positive values.
E: Structural homology models of Lp-CFTR. Lp-CFTR was modeled using the cryo-EM structures of CFTR in the closed state, left, and in the ATP-bound nearly open state, right. Domains colored as in Figure 1B. The segment of the R-domain visible in the closed state (purple) is not shown in the nearly open state.
F: Structure of the pore domain in the homology models of Lp-CFTR. Lp-CFTR was modeled using CFTR cryo-EM structures in closed (left) and ATP-bound nearly open (right) conformations, demonstrating development of a pore; residues 1101-1153 deleted in order to reveal the inner pore. Of note, F337 (L360 in Lp-CFTR, purple) occludes the pore in the closed state and S341 (S364, red) is at the narrowest spot in the pore interior although not yet rotated into the pore axis. R347 interacts with D924 (R370 and D927, upper blue-yellow pair) in both states, while R352 interacts with D993 (R375 and D996, bottom blue-yellow pair) only in the nearly open state. Of note, a key interaction between D110 in ECL1 and K892 in ECL4 observed in hCFTR in the closed state is not enabled in Lp-CFTR (N134 with R896, green pair, partially occluded in the view of the ATP-bound near-open state).
G: NBD1 of Lp-CFTR bears a key substitution. Lp-CFTR modeled using the CFTR cryo-EM structures demonstrated folding of NBD1 similar to that of human CFTR. NBD1 is noted in red ribbon and a portion of the R-domain is in purple ribbon. The lamprey substitution for phenylalanine 508, leucine 525, is noted in dashed box region and is positioned to interact with intracellular loop 4 (green ribbon).
H: NBD domains are more distant from each other in Lp-CFTR compared to the human ortholog: Comparison of distances between the centers of mass of NBD1 and NBD2 of human (black) and lamprey (red) CFTR conformations obtained from 100 ns MD simulations. (A) Closed hCFTR (5uak.pdb) and the corresponding Lp-CFTR homology model. (B) ATP-bound nearly-open hCFTR (6msm.pdb) and the corresponding Lp-CFTR homology model.
See also Data Figure S1 and Figure S1