Fig. 2. Detailed analysis of NK cell activation in COVID-19 disease.
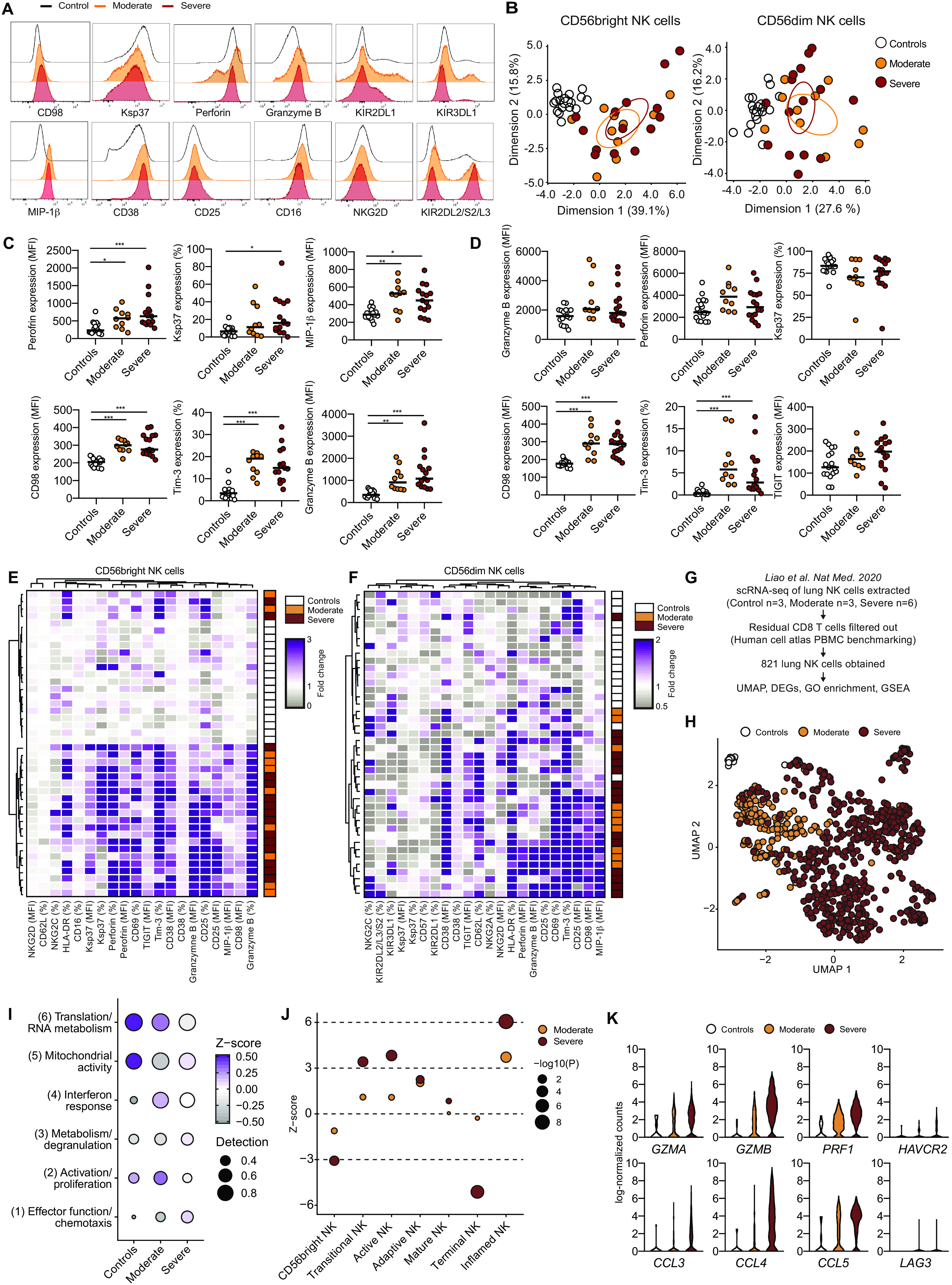
(A) Expression of indicated proteins on CD56dim NK cells. (B) PCA of the protein expression phenotype of CD56bright and CD56dim NK cells in healthy controls, moderate COVID-19 patients, and severe COVID-19 patients. (C) Expression of indicated proteins on CD56bright NK cells in healthy controls (n = 17), moderate COVID-19 patients (n = 10), and severe COVID-19 patients (n = 15). All flow cytometry measurements, including cytokines (MIP-1β and IFN-γ), were performed directly ex vivo without further stimulation. (D) Expression of indicated proteins on CD56dim NK cells in healthy controls (n = 17), moderate COVID-19 patients (n = 10), and severe COVID-19 patients (n = 16). (E and F) Hierarchical clustering heatmaps showing expression of indicated proteins compared with median of healthy controls in (E) CD56bright and (F) CD56dim NK cells. (G) Strategy for scRNA-seq analysis of BAL NK cells from controls and COVID-19 patients. (H) UMAP of scRNA-seq data for BAL NK cells from the indicated groups. (I) Heatmap of indicated gene clusters after gene ontology (GO) enrichment analysis on DEGs. (J) Z scores of NK cell gene sets after gene set enrichment analysis (GSEA). (K) Violin plots of indicated DEGs. In (C) and (D), Kruskal-Wallis test and Dunn’s multiple comparisons test; bars represent median (*P < 0.05, **P < 0.01, and ***P < 0.001).
