Figure 5.
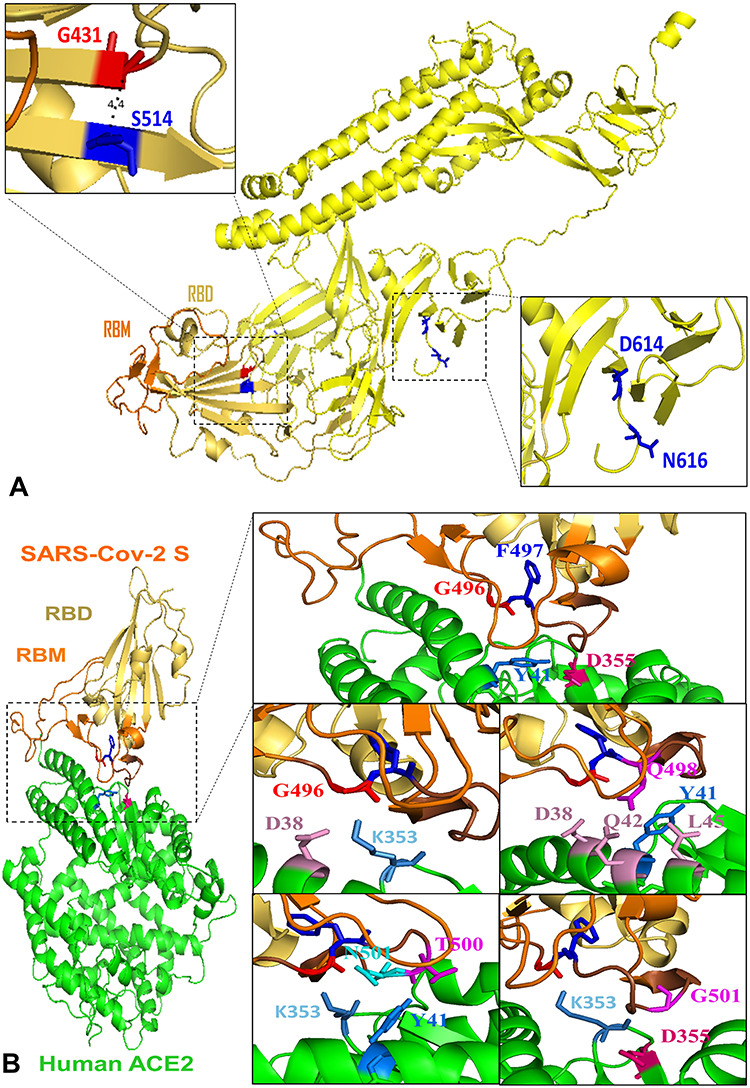
Structural representation of key residues. (A) Key residues altering protein stability in SARS-Cov-2 full-length S (yellow). G431 and K514 are spatial residues (4.4 Å) located in RBD (yellow orange) and closed to RBM (orange). Viral common variation D614G located in D614 is closed to N-linked glycosylation site N616. (B) Key residues altering the binding affinity between SARS-Cov-2 S (RBD: yellow orange; RBM: orange) and human ACE2 yellow. G496 (red) and F497 (blue) are located in the interacting motif Y495GFQPTNGVG504 (brown) and form an interaction network with ACE2 residues D355 (hot pink) and Y41 (marine). RBD residue G496 has vdw contacts with ACE2 residues D38 (pink) and K353 (sky blue). Q498 has contacts with residues D38, Q42, L45 (pink) as well as Y41 (marine). RBD residues T500 (magenta) and N501 (cyan) interact with ACE2 residues K353 (sky blue) and Y41 (marine). RBD residue G502 (magenta) has vdw contacts with D355 (hot pink) and K353 (sky blue) in ACE2.
