Figure 1.
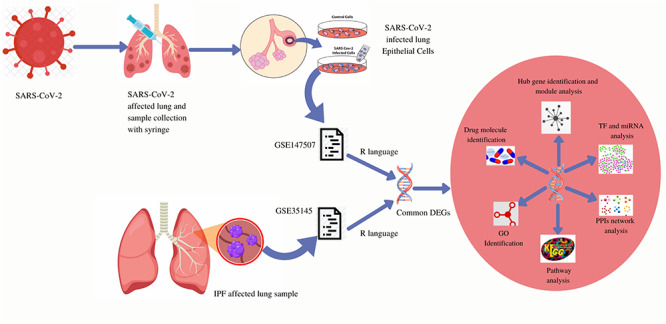
Methodical workflow for the current investigation. Two type of samples (control cells, SARS-CoV-2 infected cells) were collected from SARS-CoV-2 infected lung epithelial cells and both are included in the GSE147507 dataset. GSE147507 dataset contains a sample of SARS-CoV-2 infected lung epithelial cells and the GSE35145 dataset contains IPF affected lung samples. Common DEGs were identified from both the datasets using the R programming language. From the common DEGs, GO identification, KEGG pathway, PPIs network, TF and miRNA analysis, hub gene identification and module analysis was designed and based on those analysis drug molecule identification was performed.
