Figure 3.
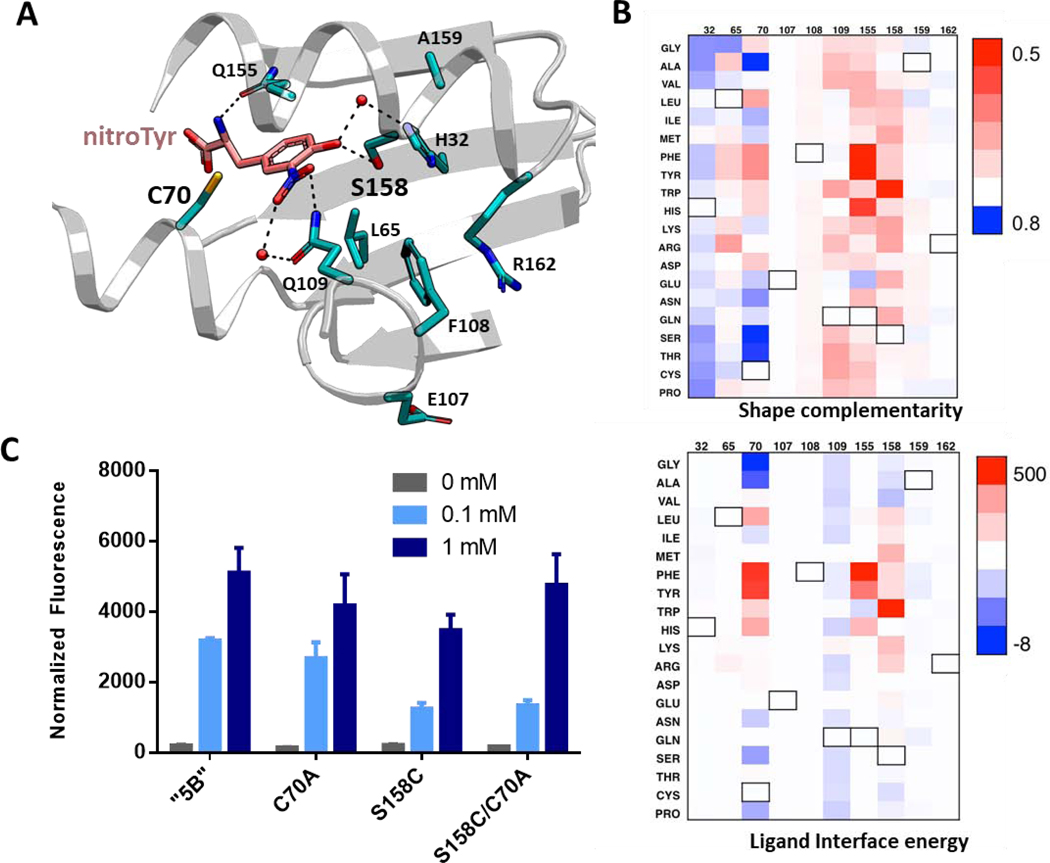
Analysis of nitroTyr binding on the active site of the “5B” aaRS modified by single residue alterations. (A) Crystal structure of the “5B” nitroTyr aaRS active site highlighting the ten residues analyzed for mutational analysis by Rosetta (PDB 4nda). (B) Analysis of shape complementarity (top) and ligand binding interface energy (bottom) for nitroTyr to each modified residue (numbers are listed on top of each panel). Black outlined boxes indicate wild-type amino acids. Value for each amino acid position is subtracted from the original value to show the absolute effect of each mutation. In both plots, blue means improved over original value. Values are provided in the Supplemental Dataset. (C) Variants of “5B” aaRS C70A, S158C and C70A/S158C predicted to have better activity were tested for their ability to express sfGFP-150TAG in vivo in RF1-containing BL21 cells by measuring in-cell fluorescence normalized to culture density. Cultures were supplemented with 0, 0.1 and 1 mM nitroTyr as indicated. Error bars represent the standard deviation of three independent cultures.
