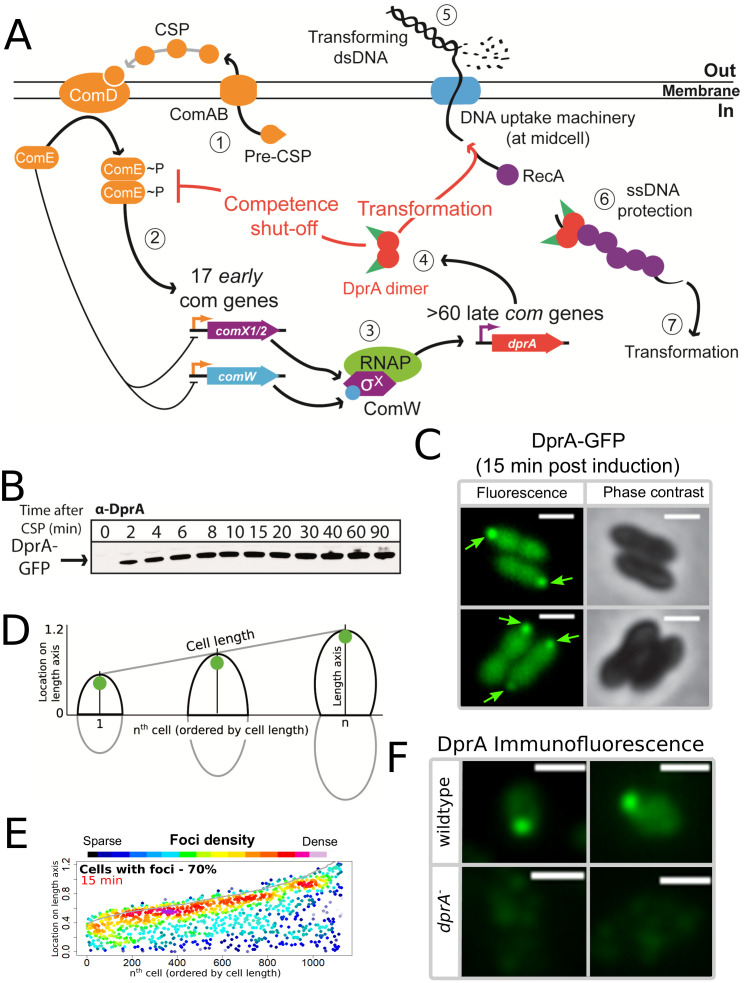
(
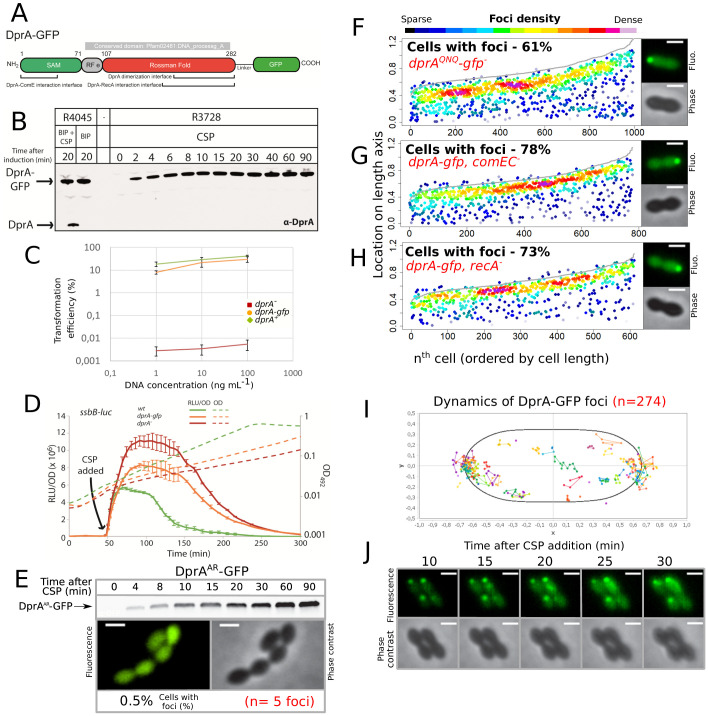
A) Linear representation of DprA-GFP with the limits of Pfam02481 (in gray) and structural domains indicated. Interfaces of dimerization and interaction with RecA and ComE are indicated and represent the regions within which the majority of point mutations affecting interaction were identified (
Mirouze et al., 2013;
Quevillon-Cheruel et al., 2012). The GFP protein is separated from DprA by a linker, as described previously (
Bergé et al., 2013). (
B) Western blot tracking cellular levels of DprA-GFP after induction as shown from strains R4045 and R3728. α-DprA antibody used. Samples at each time point corrected by OD to render direct comparison of cellular levels possible. Full gel shows no degradation of DprA-GFP throughout growth. (
C) DprA-GFP is active in transformation. Comparing transformation efficiency of wildtype (R1501),
drpA- (R2018) and
dprA-gfp (R3728) strains. R304 chromosomal DNA, conferring streptomycin resistance via
rpsL41 point mutation (
Salles et al., 1992), used to transform at 1, 10, and 100 ng mL
−1. Error bars represent triplicate repeats. (
D) DprA-GFP is partially active in competence shut-off. Comparing the competence profiles of wildtype (R1502),
drpA- (R2018) and
dprA-gfp (R3743) strains after CSP addition (100 ng mL
−1, t = 40 min). Luminometric and photometric readings of
ssbB-luc transcriptional reporter fusion taken every 5 min. Full lines represent competence induction (RLU/OD) and dotted lines represent growth (OD). Error bars represent triplicate repeats. (
E) Dimerization of DprA is necessary for accumulation of DprA-GFP at the cell poles. Western blot tracking cellular levels of DprA-GFP after competence induction in strain R4046. α-DprA antibody used. Samples at each time point corrected by OD to render direct comparison of cellular levels possible. In contrast to DprA-GFP (
Figure 1B), the levels of DprA
AR-GFP continue to increase over the time period of the experiment as competence is not shut-off in this DprA
AR strains (
Quevillon-Cheruel et al., 2012). Sample fluorescence microscopy images of R4046 strain producing DprA
AR-GFP 15 min after competence induction. Scale bars, 1 µm. 1124 cells and five foci analyzed. (
F) Disrupting the ability of DprA to interact with RecA (
dprAQNQ) does not impact the polar accumulation of DprA. Strain R4047 (
comC0, dprAQNQ-gfp) observed 15 min after competence induction. Data presented as in
Figure 1E. 767 cells and 612 foci analyzed. Sample microscopy images of strain R4047 15 min after competence induction. Scale bars, 1 µm. (
G) Inactivating the
comEC transformation pore gene to prevent uptake of exogenous DNA does not impact the polar accumulation of DprA. Strain R4082 (
dprA-gfp, comEC-) observed 15 min after competence induction. Data presented as in
Figures 1E and
6 32 cells and 778 foci analyzed. Sample microscopy images of strain R4082 15 min after competence induction. Scale bars, 1 µm. (
H) Inactivation the recombinase gene
recA to prevent homologous recombination does not impact the polar accumulation of DprA. Strain R4061 (
dprA-gfp, recA-) observed 15 min after competence induction. Data presented as in
Figures 1E and
3 93 cells and 996 foci analyzed. Sample microscopy images of strain R4061 15 min after competence induction. Scale bars, 1 µm. (
I) DprA-GFP foci remain stable over time after competence induction. DprA-GFP foci tracked by time-lapse fluorescence microscopy every 5 min from 10 to 30 min after competence induction in strain R3728. Images analyzed by MicrobeJ (
Ducret et al., 2016). Each series of linked spots represents a single focus tracked over time, localized in an averaged pneumococcal cell. (
J) Sample time-lapse fluorescence microscopy images of strain R3728 used to generate panel
H.