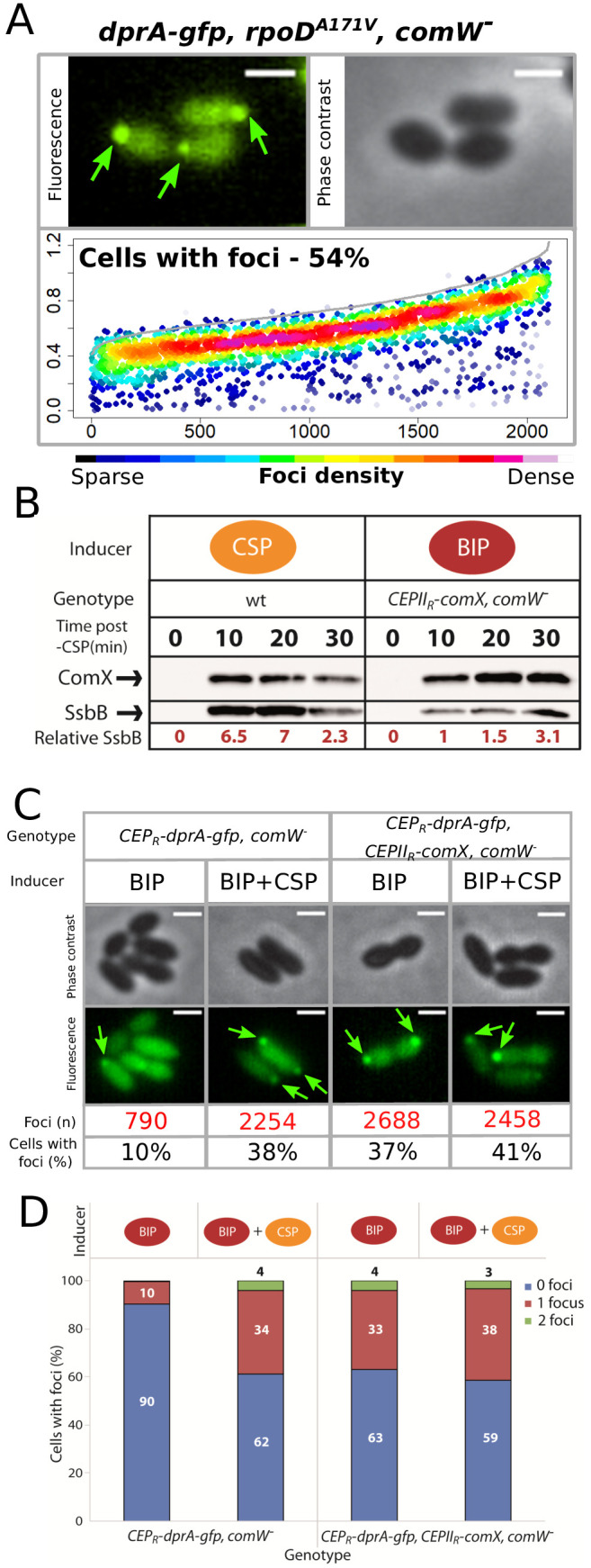
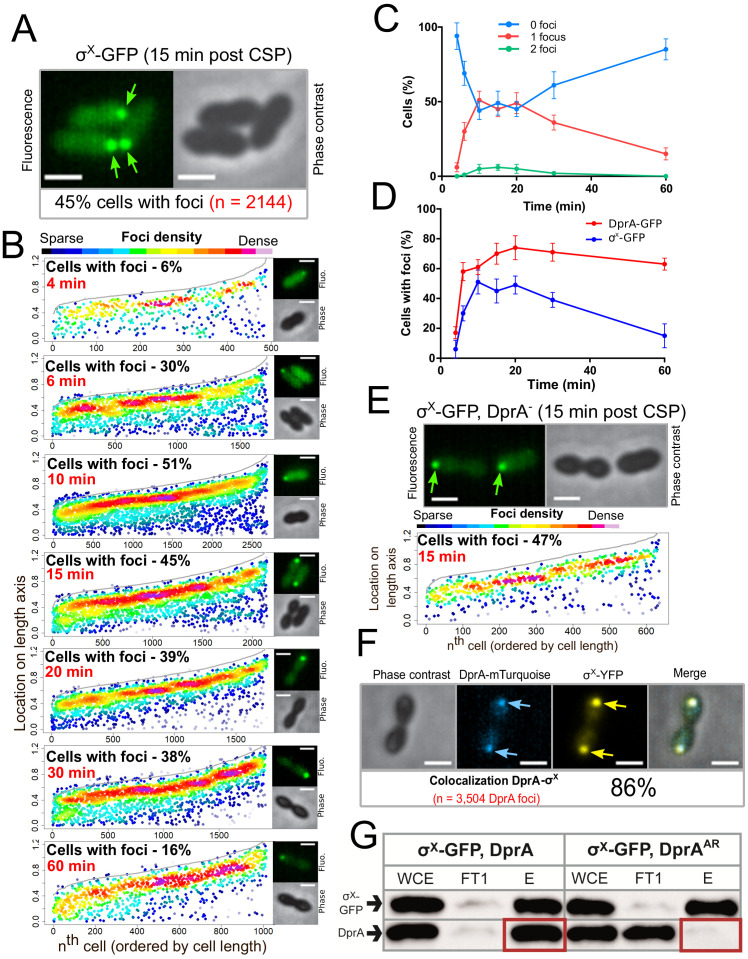
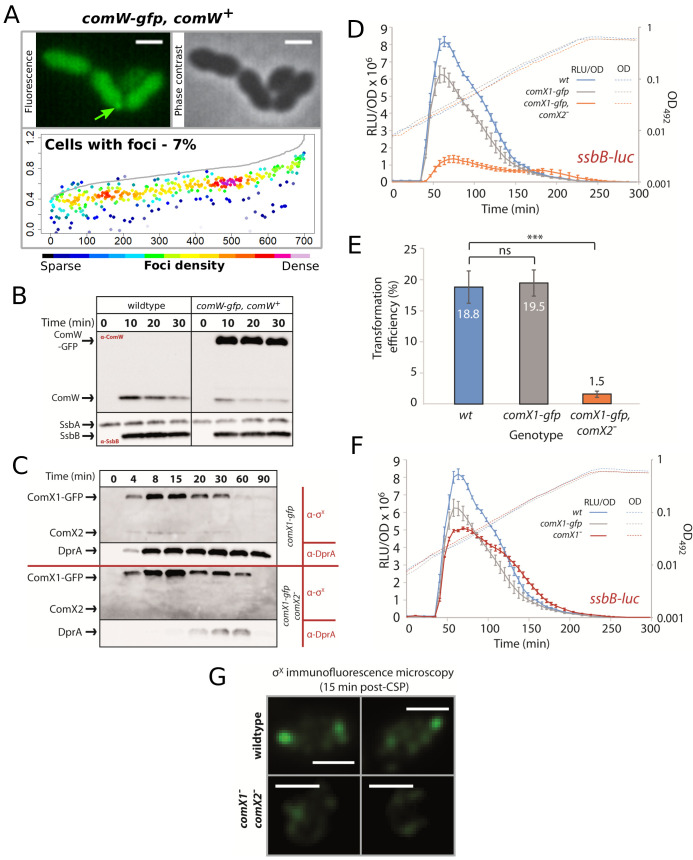
Figure 5. σX-GFP interacts directly with DprA at the cell pole during competence.
(A) Sample fluorescence microscopy images of strain R4451 producing σX-GFP from comX1 and wild-type σX from comX2 15 min after competence induction. Scale bars, 1 µm. (B) σX-GFP accumulates at the cell pole during competence. Focus density maps presented as in Figure 1E. 4 min, 7544 cells and 489 foci analyzed; 6 min, 5442 cells and 1711 foci analyzed; 10 min, 4358 cells and 2691 foci analyzed; 15 min, 3746 cells and 2144 foci analyzed; 20 min, 4211 cells and 1754 foci analyzed; 30 min, 4695 cells and 1920 foci analyzed; 60 min, 5713 cells and 1016 foci analyzed. (C) Most cells have a single σX-GFP focus. Data from the time-course experiment presented in panel B showing the number of foci per cell at each time point. Error bars represent triplicate repeats. (D) DprA-GFP foci persist in cells longer than σX-GFP foci. Comparison of cells with foci at different timepoints from timecourse experiments. DprA-GFP from Figure 3A, σX-GFP from panel B. Error bars represent triplicate repeats. (E) Accumulation of σX at the cell poles does not depend on DprA. Sample microscopy images of a comX1-gfp, dprA- strain (R4469). Focus density maps generated from cells visualized 15 min after competence induction presented as in Figure 1E. 1104 cells and 638 foci analyzed. (F) σX and DprA colocalize at the cell pole. Colocalization of σX-YFP and DprA-mTurquoise in R4473 cells visualized 15 min after competence induction. 7460 cells and 3504 DprA-mTurquoise foci analyzed. Scale bars, 1 µm. (G) DprA is copurified with σX-GFP while DprAAR is not. Western blot of pull-down experiment carried out on strains producing σX-GFP and either DprA (R4451) or DprAAR (R4514) 10 min after competence induction. WCE, whole cell extract; FT1, flow through; E, eluate.
Figure 5—figure supplement 1. σX is necessary and sufficient to mediate accumulation of DprA at the cell poles.