Figure 4.
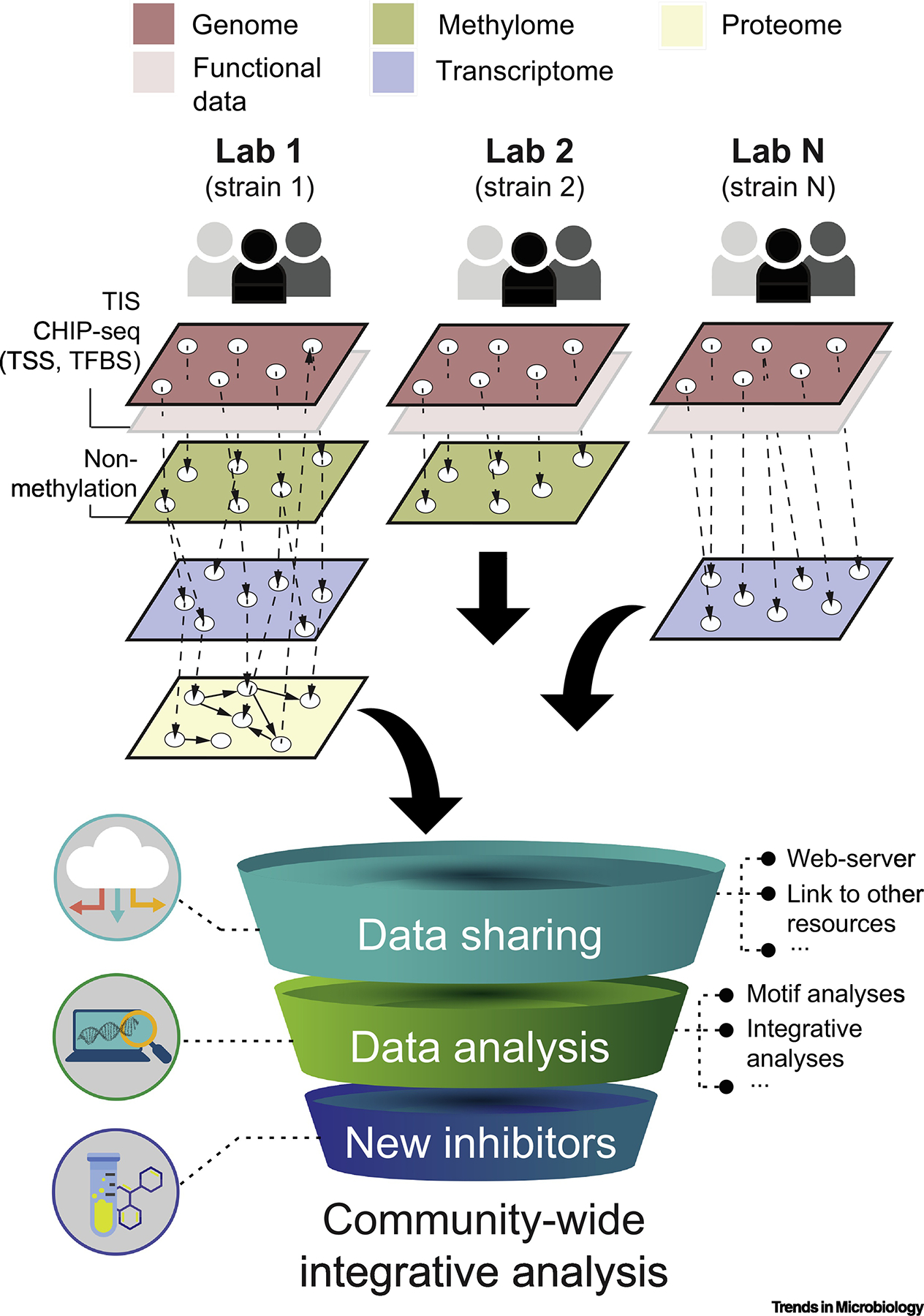
Overview of a large-scale community-wide integrative approach for bacterial methylome analyses. Core and persistent MTases can be prioritized to build a trans-omic network across multiple laboratories merging multiple functional data gathered at different experimental conditions. The latter may build upon MTase mutants generated by Transposon Insertion mutagenesis coupled with deep Sequencing (TIS), site-directed mutagenesis of methylation sites, genome-wide profiling of DNA binding proteins (ChIP-seq), transcription start site (TSS) mapping, and identification of methylation-sensitive transcription factors. Multiple layers of omics data may ultimately be commonly shared, linked to other resources (e.g., REBASE), allow for an in-depth analysis of, for example, methylation motif conservation, phase-variable DNA methyltransferases, and accelerate the research of novel epigenomic inhibitors.
