Figure 4. Structural Analysis of Fab RRV-12 Binding to RRV, CHIKV, and MAYV by Cryo-EM.
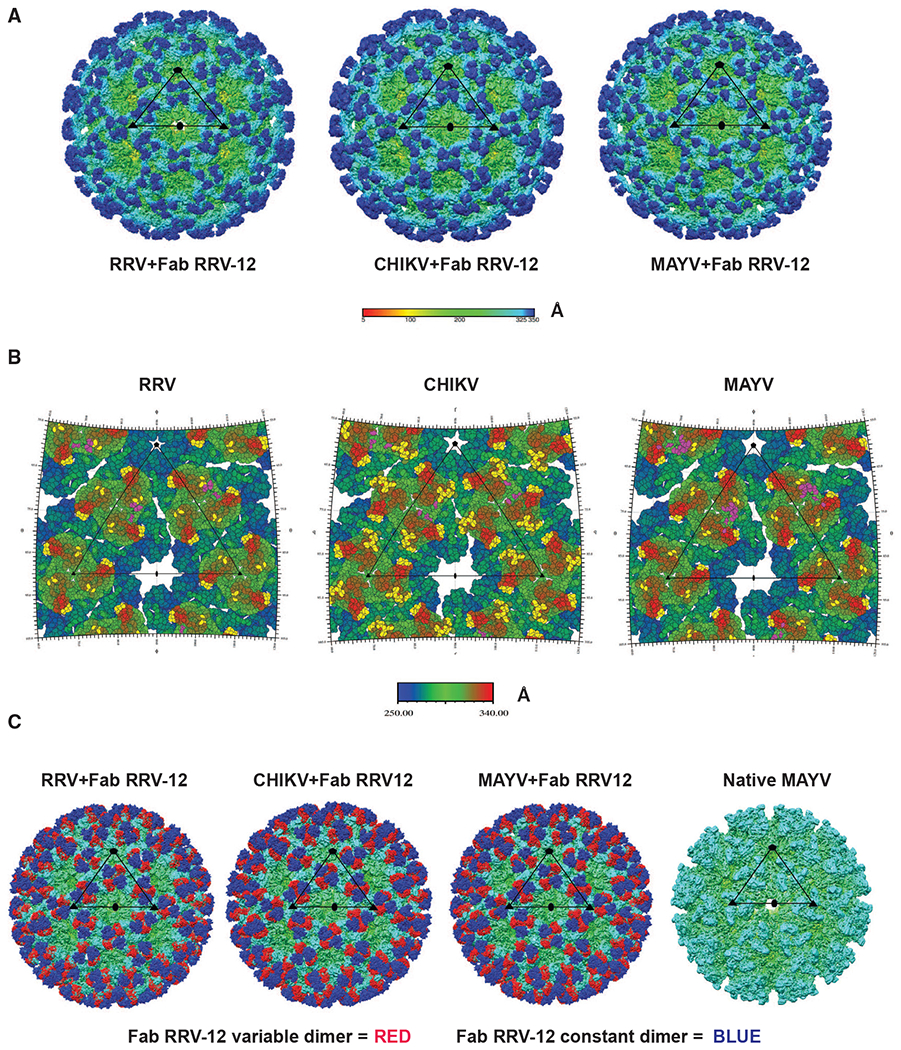
(A) Cryo-EM density maps of RRV, CHIKV, and MAYV bound by Fab RRV-12. Each map was determined by single-particle reconstruction and icosahedral averaging. The scale bar represents the radial distance from the particle center by color in angstroms. The superimposed black triangle represents the asymmetric unit; symmetry axes are indicated by a pentagon for the 5-fold axis, a triangle for the icosahedral 3-fold axis, and an oval for the 2-fold axis. The resolution of each map is 6.3Å for RRV and 5.3Å for both CHIKV and MAYV.
(B) RIVEM road maps of the viral surface. Scale bar, Å, is radial distance from the center of the virus. Residues highlighted in yellow on RRV and MAYV E2 B domain, E2 A domain, and β-ribbon, and CHIKV E2 B domain and E3 and are all located within 6 Å or less to the backbone of the fitted Fab structure. Highlighted residues indicate the B domain of each i3 and q3 trimer is fully occupied. Residues highlighted in purple indicate residues of the viral surface 6 Å or less to the position of the backbone structures of Mxra8. Mxra8 occupies only one position per trimer.
(C) Cryo-density maps of RRV, CHIKV, and MAYV bound with Fab RRV-12 and native MAYV structure. Fab constant or variable domains are colored blue or red, respectively. The antibody variable domain binds the B domain of one trimer and the antibody constant domain extends to cover an adjacent trimer. The native structure is shown for a comparison with an unoccupied trimer. The remaining density of each structure is colored based on radial distance from the center of the particle, see scale bar in (Å).
