Figure 5. Fab RRV-12 Variable Domain Binds the B Domain.
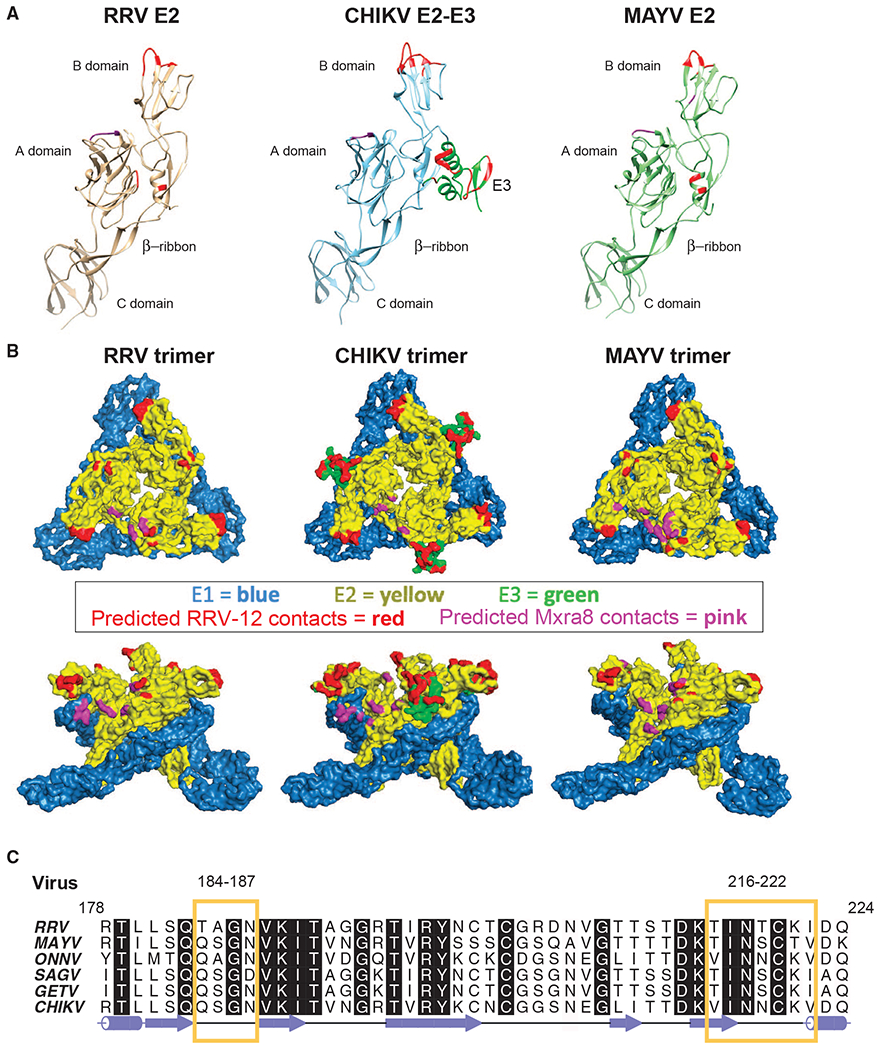
(A) Ribbon diagrams of E2 ectodomain structure from each virus. Regions of the B domain highlighted in red are the predicted epitope. Highlighted regions span residues RRV 183–187, 218–221, and 223; CHIKV E2 179–184, 198–200, and 213–219; and MAYV E2 184–187 and 219–221. Residues highlighted in red on the RRV and MAYV A domain and β-ribbon and on CHIKV E3 are predicted to be in close vicinity of the constant domain due to orientation of the Fab to the viral surface. Residues RRV 25–28 and 61–63, CHIKV 22–24 and 192, and MAYV 25–27 and 192, shown in purple, are predicted to be the Mxra8 binding site.
(B) Surface representations of RRV, CHIKV, and MAYV trimer generated from the fitted asymmetric unit. E1 and E2 are blue and yellow, respectively. E3 on the CHIKV trimer is shown in green. The surface region highlighted in red on the B domain of each trimer is the predicted epitope. Positions highlighted in red on the RRV and MAYV A domain and β-ribbon are part of the constant domain footprint. Surfaces of CHIKV E3 highlighted in red are part of both the variable and constant domain footprint. Areas highlighted in purple are the predicted position of Mxra8.
(C) Amino acid sequence alignment of E2 B domain residues 178–224 from viruses CHIKV, ONNV, MAYV, RRV, SAGV, GETV, generated using ALINE program (Bond and Schüttelkopf, 2009). Yellow boxes outline semi-conserved regions between the viruses containing residues of the Fab RRV-12 epitope.
