Figure 1: Experimental strategies for leveraging genetic tractability and diversity to understand complex phenotypes.
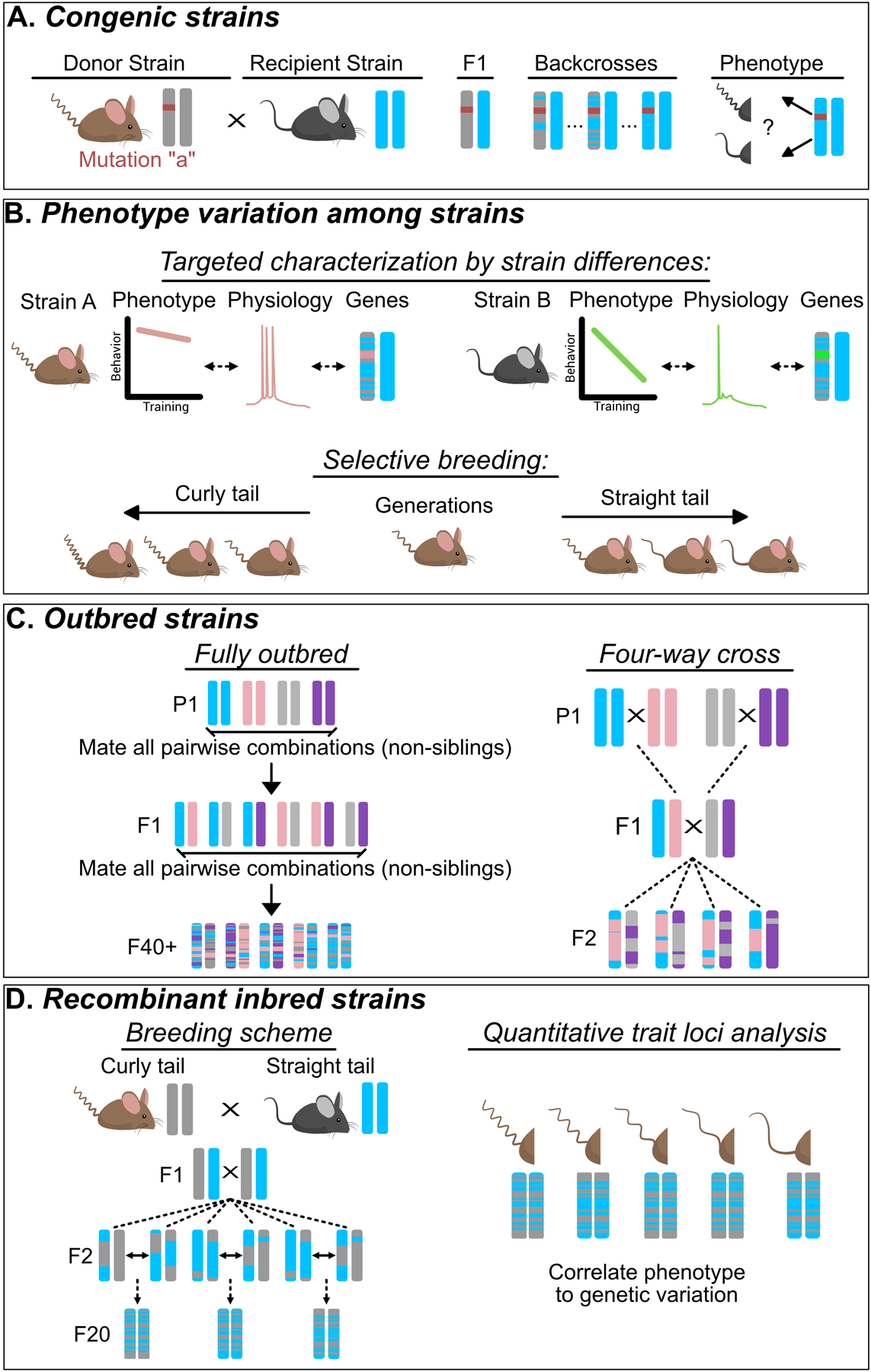
While these strategies can be used to study complex behavior, the physical trait of a curly tail has been used for this illustration. (A) Congenic Strains are used to test for the presence of gene modifiers on a phenotype of interest by determining if the phenotype is maintained in both the donor and recipient strain. (B) Phenotype variation among strains. Targeted characterization by strain differences (top) can be used to link physiological differences among strains to differential phenotypes. When differences between stains are not natively present, selective breeding (bottom) can be employed to generate quantitative divergence for a trait. (C) Outbred strains represent an important technique for testing hypotheses that rely on genetic heterogeneity, such as validating the generalizability of experimental treatments. Distinct outbred breeding strategies yield differences in the amount of genetic diversity and reproducibility. For example, a fully outbred scheme has higher diversity, but lower reproducibility, while a four-way cross has lower diversity, but higher reproducibility. Note that the more unique parental (P1) strains used, the more genetic diversity is generated (D) Recombinant inbred strains have the highest amount of genetic diversity while preserving genetic tractability. Typically, they are used for quantitative trait locus analysis (QTL) which is a technique that matches variation in gene expression to variation in a quantitative trait or phenotype.
