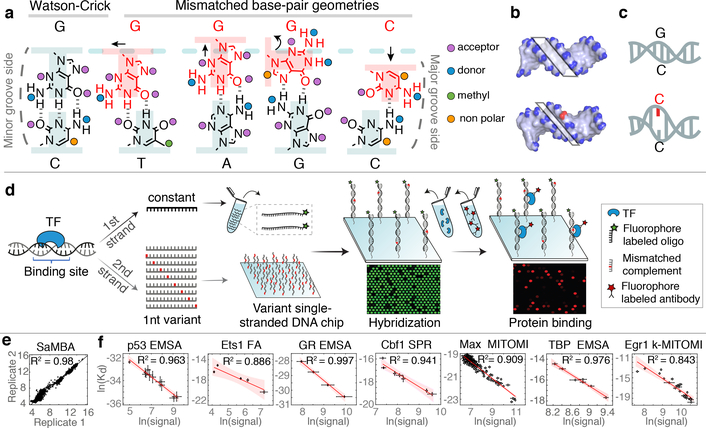
Figure 1. SaMBA measures the effects of mismatches on protein-DNA binding in high throughput.
(a-c) Mismatches change the local DNA geometry (a), affect global features such as the minor groove width (b), and destabilize the DNA (c).
(d) SaMBA is a chip-based assay for testing TF binding to thousands of DNA mismatches and Watson-Crick sequences (Methods). DNA hybridization and protein-DNA binding are quantified using fluorophore-labeled oligos and antibodies, respectively.
(e) Reproducibility of SaMBA data, for technical replicates of Ets1 at 125nM. Axes show the base 2 logarithm of the median fluorescent intensity signal corresponding to the bound Ets1 protein (n=12 replicate spots for Watson-Crick sequence, and 8 for mismatched sequences).
(f) Protein binding levels measured by SaMBA correlate linearly with independent Kd measurements from a variety of experimental methods, allowing calibration of SaMBA data. Similarly to related array-based techniques20, median values over replicate DNA spots are shown for SaMBA (error bars: median absolute deviation). Average values over replicates are shown for the orthogonal methods (error bars: standard deviation, when available). See Methods for the number of replicates (n>=3) for each experiment. Red shaded region: 95% confidence interval for Pearson’s correlation.