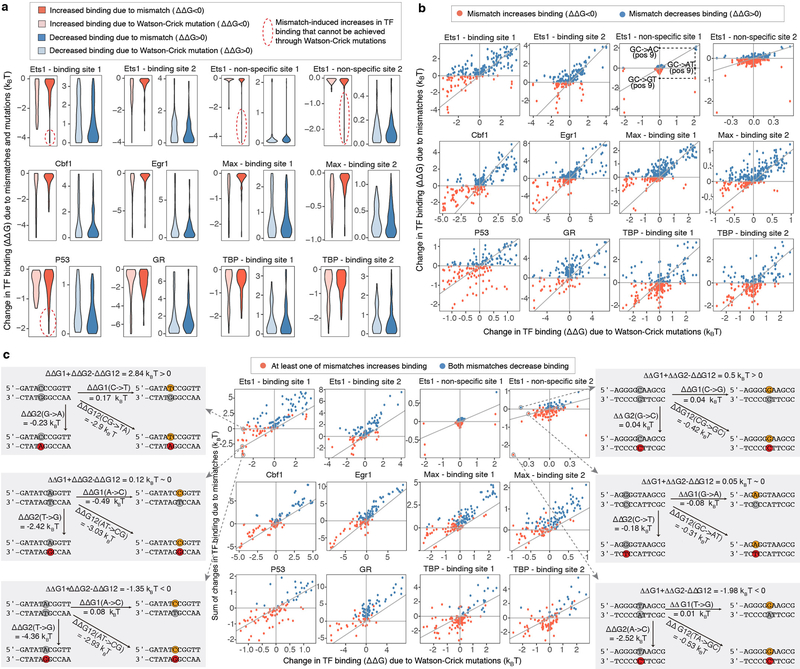
Extended Data Figure 4. Comparing the effects of mutations versus mismatches on TF binding.
(a) The magnitude of the energetic effects of mutations (light colors) and mismatches (dark colors) is similar. The effects were computed for all 7 proteins with available calibration data in our study, and for a total of 12 DNA sites (Methods). The effects of mismatches were calculated relative to the two closest Watson-Crick sequences (e.g. for a G-T mismatch the closest Watson-Crick base pairs are G-C and A-T; the mismatch plots include both ΔΔG(G-C -> G-T) and ΔΔG(A-T -> G-T)).
(b) Mismatches and their corresponding mutations have different, even opposite effects on TF binding. Each mutation is compared to the two closest mismatches (e.g. G-C -> A-T is compared to both G-C -> A-C and G-C -> G-T). Upper left quadrant: mutations increase binding, mismatches decrease binding. Upper right quadrant: both mutations and mismatches decrease binding. Lower left quadrant: both mutations and mismatches increase binding. Lower right quadrant: mutations decrease binding, mismatches increase binding. X-axis and Y-axis show calibrated binding measurements computed from the median SaMBA signal intensities (over n=10 replicate spots).
(c) Comparing the effect of mutations versus the cumulative effects of the two closest mismatches. Points close to the diagonal correspond to cases where the effect of the mutation is approximately equal (within experimental noise) to the sum of the effects of the two mismatches. Points above the diagonal correspond to cases where Watson-Crick mutations have either a more beneficial or a less detrimental effect on TF binding compared to the cumulative effect of the two mismatches. Points below the diagonal correspond to cases where Watson-Crick mutations have either a less beneficial or a more detrimental effect on TF binding compared to the cumulative effect of the two mismatches. X-axis and Y-axis show calibrated binding measurements computed from the median SaMBA signal intensities (over n=10 replicate spots). Please see Supplementary Table 4 for the raw binding data used to compute the measurements shown in this figure.