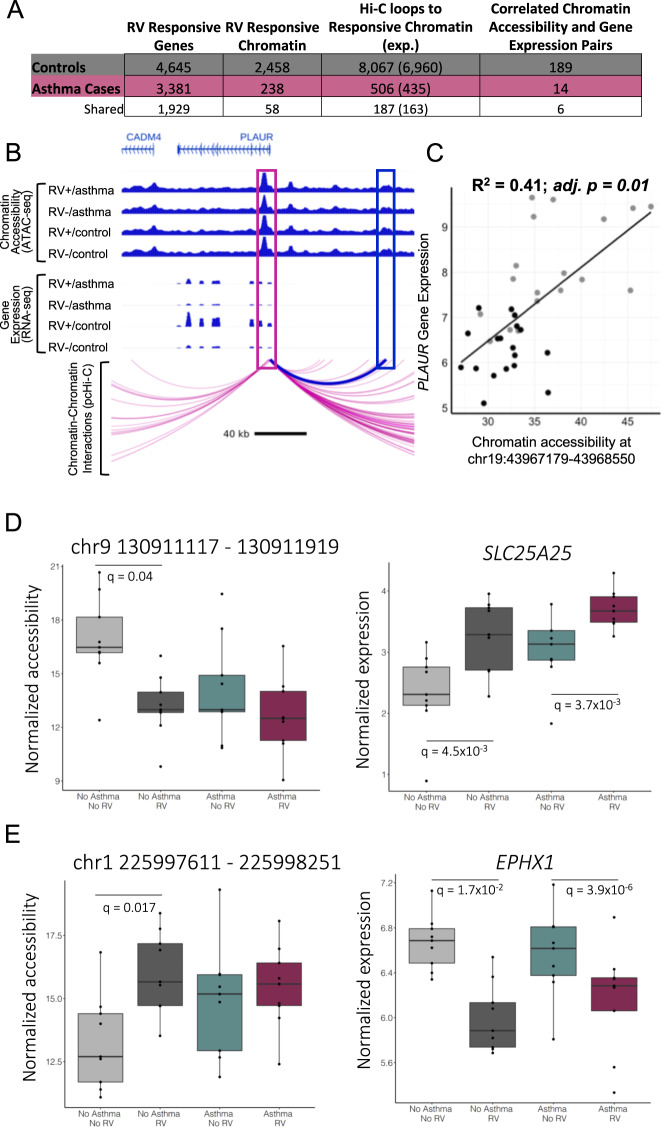
Fig. 5. RV-responsive areas of open chromatin correlated with gene expression at pcHi-C predicted target genes.
A The numbers of RV-responsive genes, chromatin regions, or pcHi-C target genes of RV-responsive chromatin regions in controls (gray) and cases (burgundy) and both (white) are shown in the first three columns. The last column shows the number the pcHi-C predicted genes where the chromatin accessible and gene expression were correlated (Spearman’s correlation, FDR adj. p < 0.05). B The promoter of PLAUR loops to an area of open chromatin at chr19:43967179–43968550. Chromatin accessibility for each condition is shown in the top panel, gene expression in the second panel and the bottom panel indicates pcHi-C looping from the PLAUR promoter. The pink box indicates the PLAUR promoter, the blue box indicates the RV-responsive area of open chromatin. C Correlation between expression of PLAUR and chromatin accessibility at chr19:43967179–43968550. D, E are gene-chromatin pairs where the gene is RV-responsive in both cases and controls while the chromatin is only RV-responsive in the controls.