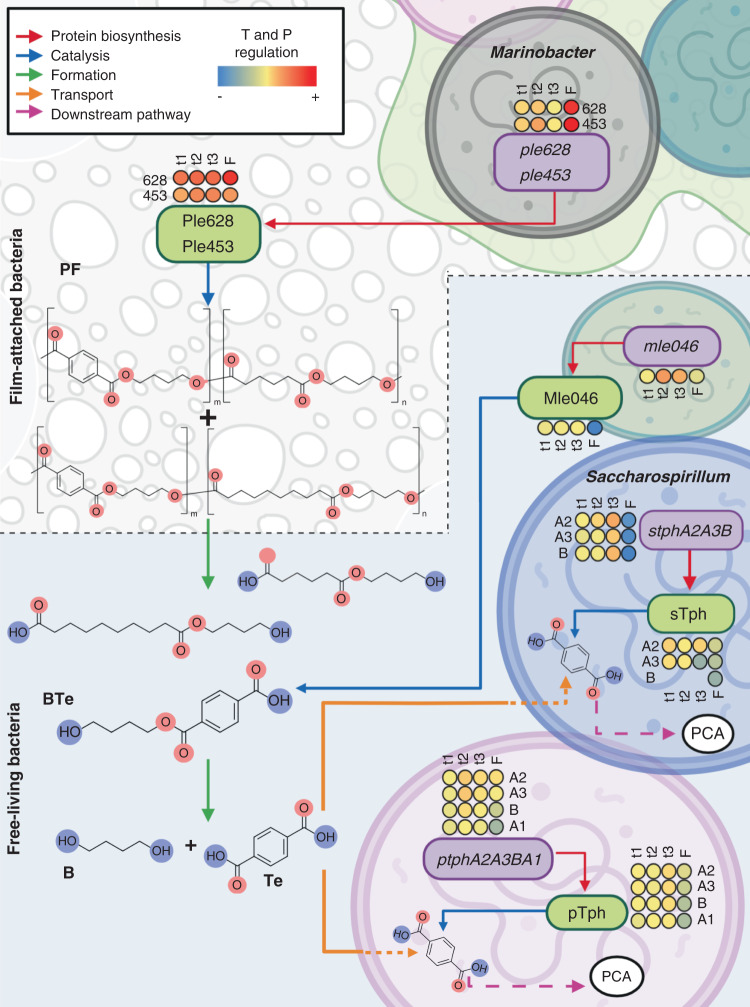
Fig. 6. Integration of multi-omics data.
Proposed mechanisms for the synergistic biodegradation of a PBAT-based blend film (PF) during different time points (t1–t3) and within a biofilm (F). Periplasmatic α/β hydrolases (Ple628, Ple453), synthesized by Marinobacter, hydrolyze PF to produce oligomers and monoesters of 1,4-butanediol (B) with either terephthalic acid (Te) or sebacic acid (Se)/adipic acid (Ad). An Alphaproteobacterial Mle (Mle046) hydrolyzes a mixture of terephthalate-butanediol monoester (BTe) to produce Te and B. Te is transported into the cell and catabolized by terephthalate degradation clusters of an unknown bacterium (pTPD), and to a lesser extent by Saccharospirillum (sTPD). Downstream degrading genes leads to the formation of protocatechuate (PCA). Red arrows indicate gene translation into a functional protein; blue arrows indicate the catalysis of a substrate by a protein; green arrows, show the formation of intermediates by the catalysis of a substrate; orange arrows represents the transport of intermediates from the surroundings to bacteria; pink arrows represent donwstream degradation pathways. The color scale represent upregulated (+) and downregulated (−) transcripts (T) and proteins (P) (created with Biorender.com).