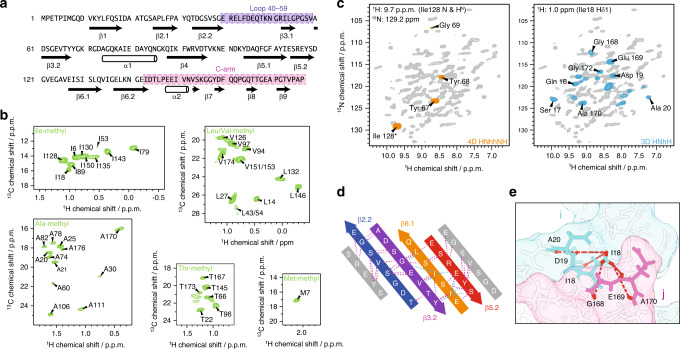
Fig. 1. Solid-state NMR data for hybrid structure calculation defines local structure.
a Backbone dihedral angles define the secondary structure (arrows represent β-sheets, barrels α-helices). The loop (40–59) and the C-arm (143–176) are highlighted in purple and pink, respectively. b Isoleucine Cδ1-methyl, alanine Cβ-methyl, leucine/valine Cδ/Cγ-methyl, threonine Cγ2-methyl, and methionine Cε-methyl labeling renders those moieties NMR-visible and yields highly resolved NMR spectra (green). c Long-range restraints between amide protons are extracted from a 4D HNhhNH spectrum (orange), whereas long-range restraints between amide and methyl groups are extracted from a series of 3D HNhH spectra (blue). 2D planes from both types of spectra are superimposed on a 2D hNH fingerprint spectrum (gray). d Representative long-range restraints (dashed lines) defining the inner β-barrel of the tail tube. The colored β-strands belong to one monomer, whereas the gray β-strands are from the neighboring subunits. Amide–amide contacts are highlighted in violet, amide–methyl contacts in cyan. e Schematic representation of long-range distance restraints between the Cδ1 methyl group of Ile18 and amide groups, defining the interface between the N-terminus of one monomer i (including Ile18; in cyan) and the C-arm (143–176) of another monomer j (in pink) within the tail tube. Protons are colored in red.