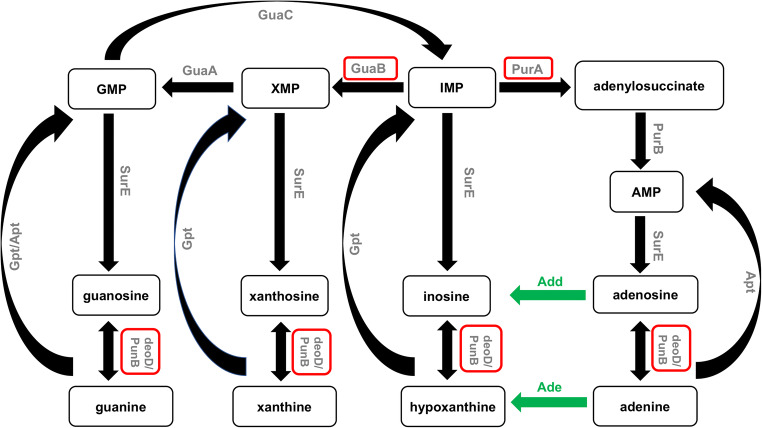
Fig. 2.
The purine nucleotide biosynthesis pathway in H. pylori. Enzymes that have been studied in H. pylori by mutant analysis and/or biochemical analysis are shown in gray. Enzymes described in this work are shown in red frames. Enzymes with likely functions, whose genes have not yet been identified, are shown in green (figure adapted from Liechti and Goldberg (2012) and Miller et al. (2012)). Abbreviations: GuaB, IMP dehydrogenase; GuaA, GMP synthetase; GuaC, GMP reductase; PurA, adenylosuccinate synthetase; PurB, adenylosuccinate lyase; Gpt, hypoxanthine-guanine phosphoribosyl-transferase; Apt, adenine phosphoribosyltransferase; SurE, 5′-nucleotidase; deoD, gene encoding purine nucleoside phosphorylase; PunB, purine nucleoside phosphorylase; Ade, adenine deaminase; Add, adenosine deaminase; IMP, inosine monophosphate; XMP, xanthosine monophosphate; GMP, guanosine monophosphate; AMP, adenosine monophosphate