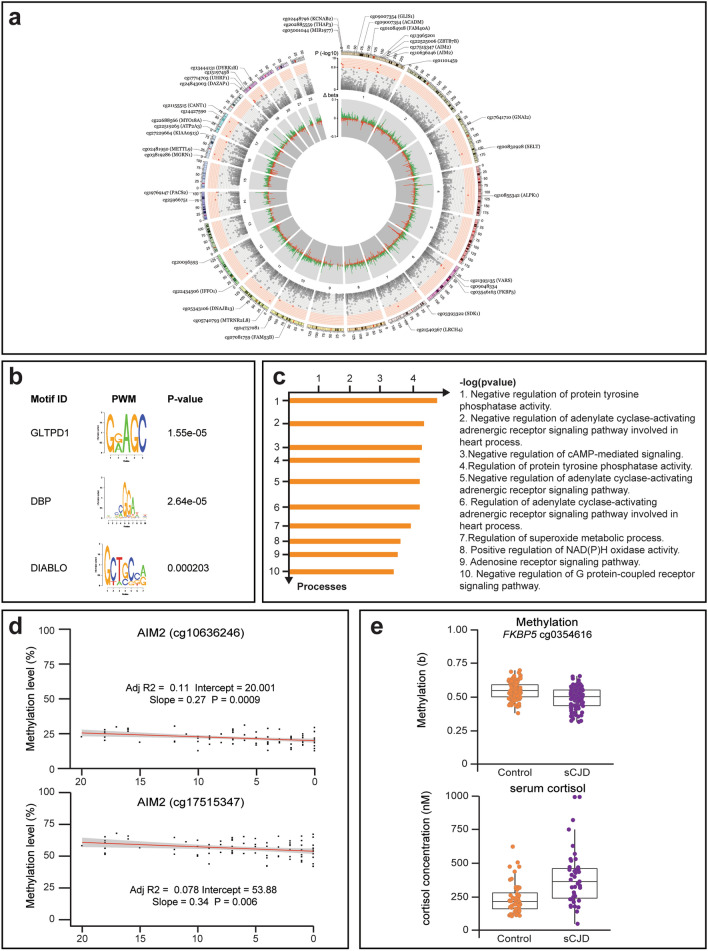
Fig. 2.
Key findings replicated using pyrosequencing in an independent case–control cohort. a Circos plot of epigenome-wide methylation levels in sCJD. Outermost circle represents the chromosome ideogram. Middle circle shows p values (− log10) of the top 25,000 most significant DMPs (Bonferroni significance threshold of p < 1.24 × 10–7 or − log10(p) > 6.9 is distinguished with a red background). Significant DMPs are shown in red and labelled accordingly. The innermost circle represents the ∆β values across the genome, with hypermethylation in green and hypomethylation in red. b Top ten motifs enriched in sequences within ± 122 bases flanking the CG found under ‘Forward_Sequence’ heading in the Illumina HumanMethylation450 manifest file of the 38 significant DMPs. Ranking based on p value. c Top ten gene ontologies enriched in genes overlapping DMPs as identified using MetaCore (p value threshold = 0.1). d Pearson coefficients between MRC Scale score with hypomethylation at two CpGs in the AIM2 promoter (cg10636246 and cg17515347). e Left: DNA methylation levels (%) at FKBP5 cg03546163 from the discovery study in sCJD patients (n = 114; purple) and healthy controls (n = 105; orange) limma p = 1.07e−07, corrected p = 0.043. Right: serum cortisol concentrations (nM) in 39 sCJD patients (purple, 239.8 nM) and 52 controls (orange, 387.6 nM) (p = 6.6 × 10–5)