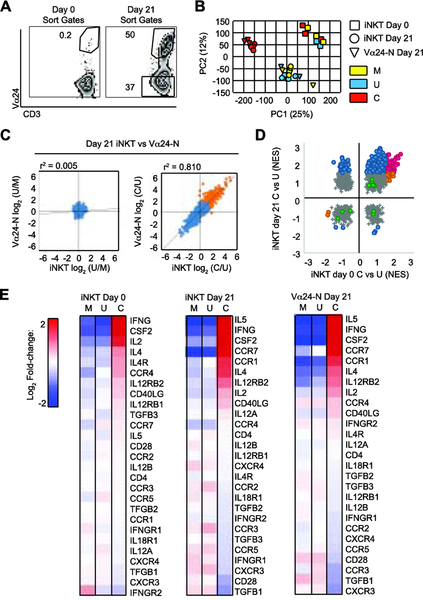
Figure 4: Expanded iNKT cells and Vα24-N cells share gene expression profiles.
(A) Representative gates used to sort day 0 CD3+Vα24+ iNKT cells (left panel) and day 21 (right panel) CD3+Vα24+ iNKT cell and CD3+Vα24neg Vα24-N cell populations.
(B) Principal component analysis (PCA) from gene expression of sorted day 0 iNKT cells (squares) (N = 3 per condition), day 21 iNKT cells (circles) (N = 4 per condition), and Vα24-N cells (triangles) (N = 4 per condition) stimulated 24 hours with media control (Stim M, yellow), unloaded bead control (Stim U, blue), or anti-CD2/CD3/CD28-loaded beads (Stim C, red).
(C) Mean fold gene upregulation (log2) over mean background in day 21 iNKT cells vs Vα24-N cells given Stim C vs Stim U (C/U, day 21) (right panel) and Stim U vs Stim M (U/M, day 21) (left panel). Stimulations are as defined in B. Correlation coefficient (r2) is indicated above each panel.
(D) Normalized enrichment score (NES) for pathway activity identified by gene set enrichment analysis (GSEA) in sorted day 0 iNKT cells given Stim C vs Stim U (iNKT C/U, day 0) vs day21 iNKT cells given Stim C vs Stim U (iNKT C/U, day 21). Stimulations are as defined in B. Blue, pathways significantly up- or down-regulated at day 21 but not at day 0; orange, pathways significantly up- or down-regulated at day 0 but not at day 21; magenta, pathways significantly up- or down-regulated at both day 0 and day 21; grey, pathways not significantly altered at day 0 or day 21.; and green, conventional T cell activation pathways,
(E) Biocarta NKT pathway gene expression profile. Heat map of log2 signal change in day 0 iNKT cells (N = 3), day 21 iNKT cells (N = 4), and Vα24-N cells (N = 4) stimulated under conditions identical to those in B. Red, increased relative expression; blue, decreased relative expression.