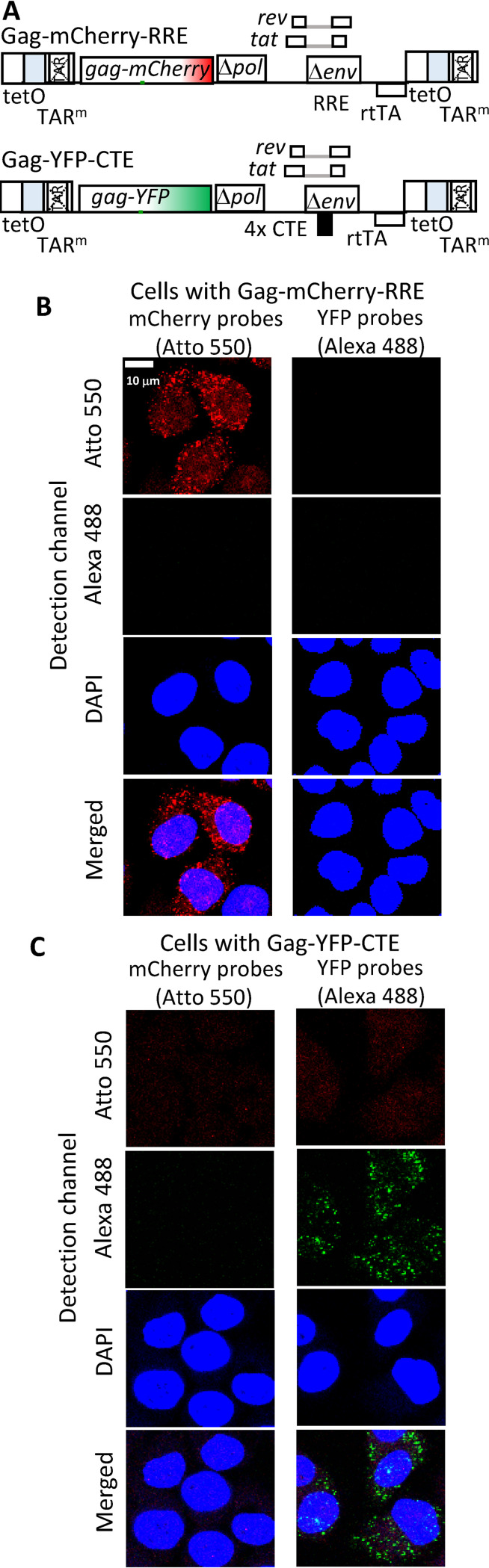
FIG 4.
Experimental system used to study expression kinetics and cytoplasmic locations of HIV-1 RNA exported via the CRM1 or NXF1 pathway. (A) General structures of inducible HIV-1 constructs. In these constructs, the rtTA gene was inserted into the nef gene, and the tet operator (tetO) binding sites were inserted into the HIV-1 LTR. (B and C) Representative images of RNAscope detection of HIV-1 RNA derived from Gag-mCherry-RRE provirus (B) or Gag-YFP-CTE provirus (C). Probes specific to the mCherry or yfp gene used in the experiments are indicated above the panels. Channels used to detect signals are shown on the side of the images. Signals from mCherry probes are shown in red, whereas signals from yfp probes are shown in green. DAPI stains are shown in blue.