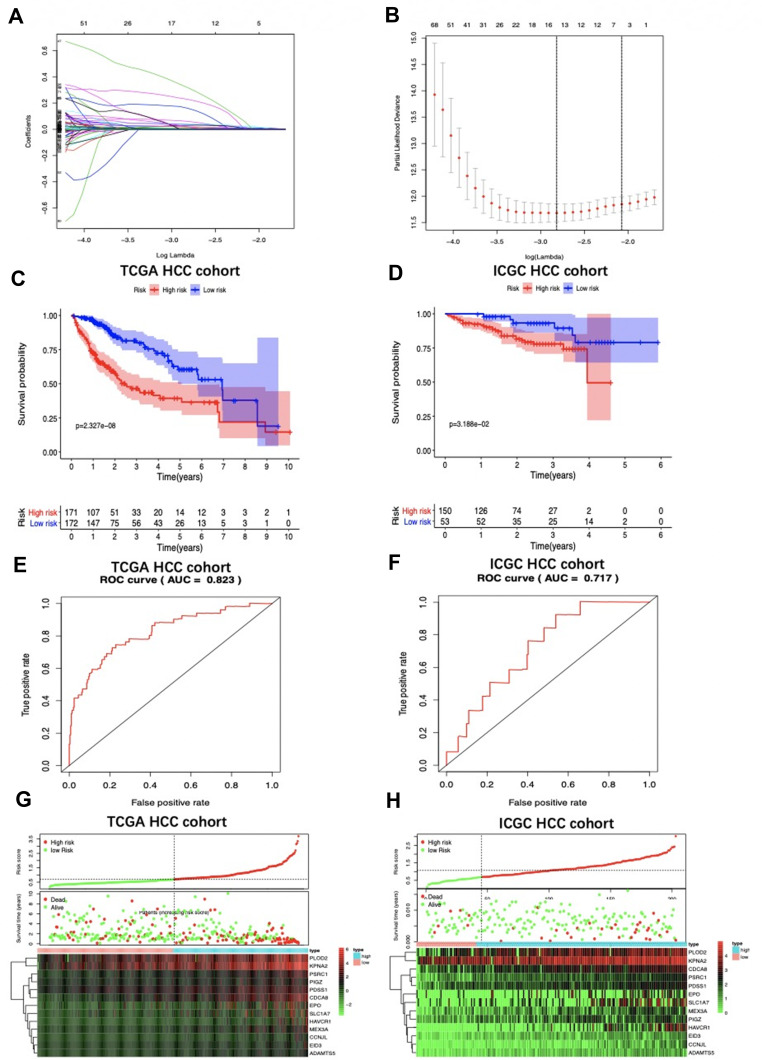
Figure 3.
LASSO coefficient profiles are depicted in (A). (B) shows the selection of the tuning parameter (lambda) in the LASSO model by tenfold cross-validation based on minimum criteria for OS; the lower X axis shows log (lambda), and the upper X axis shows the average number of OS-genes. The Y axis indicates partial likelihood deviance error. Red dots represent average partial likelihood deviances for every model with a given lambda, and vertical bars indicate the upper and lower values of the partial likelihood deviance errors. The vertical black dotted lines define the optimal values of lambda, which provides the best fit. Survival curves of patients in high risk group and low risk group of The Cancer Genome Atlas (TCGA) hepatocellular carcinoma (HCC) cohort (C) and the International Cancer Genome Consortium database (ICGC) HCC cohort (D) are exhibited. Patients in high-risk group suffered shorter overall survival. (E) and (F) show survival-dependent receiver operating characteristic (ROC) curves validation at 1 – year of prognostic value of the prognostic index in the two databases (TCGA and ICGC, respectively). Distribution of risk score, overall survival (OS), gene expression in (G) TCGA and (H) ICGC databases were also shown. Distribution of risk score, OS and heat map of the expression of eleven genes in low-risk and high-risk groups are listed in the picture from top to bottom.