Abstract
Background
Pinus massoniana Lamb. is an important afforestation tree species with high economic, ecological and medicinal values. Aluminum (Al) toxicity driven by soil acidification causes dieback of P. massoniana plantations. Previous studies showed that ectomycorrhizal fungi alleviate Al stress damages in Pinus, but the underlying molecular mechanisms and key genes induced by ectomycorrhizal fungi inoculation under Al stress in Pinus have not been explored. Herein, we applied Al stress for 60 days to P. massoniana seedlings inoculated with Suillus luteus (SL) and those non-inoculated. Then, we compared their growth parameters and transcriptome in order to detect candidate genes induced by SL conferring Al tolerance in P. massoniana.
Result
Our results showed that SL inoculation confers Al stress tolerance in P. massoniana through improved growth performance, strong antioxidant enzyme activities and reduced malondialdehyde accumulation as compared to non-inoculated seedlings. Transcriptome sequencing further supported these findings as very few genes (51 genes) were transcriptionally altered by Al in SL inoculated plants as compared to non-inoculated plants (2140 genes). We identified three core genes (cox1, cox3 and Nd1) that were strongly up-regulated by Al in the SL inoculated plants but were down-regulated in the non-inoculated plants. We also identified 42 genes specifically regulated by SL inoculated plants under Al stress, which are involved in a wide range of biological processes such as antioxidative response, transporters, hormone signaling and plant pathogen infection responses.
Conclusions
Altogether, our data suggest that SL inoculation induces priming of key stress response pathways and triggers specific genes that efficiently alleviate Al stress effects in P. massoniana. The candidate genes resources generated in this study are of utmost importance for functional characterization and molecular studies aiming at improving Al tolerance in plants.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-020-02719-3.
Keywords: Aluminum toxicity, Afforestation, Gene expression, Ectomycorrhizal fungus, Pinus massoniana
Background
Pinus massoniana Lamb. is an important afforestation tree species belonging to the Pinaceae family. It is native to southern China and is one of the dominant species for forest plantations in China [1]. P. massoniana is a pioneer species, highly tolerant to environmental stresses and grows well in barren areas and metal-contaminated soils [2–6]. It not only contributes to meeting the growing demand of wood products but also reduces pressures on natural forests and significantly contributes to restoration of degraded soils [7–10]. Besides these economic and ecological values, several studies have demonstrated the pharmacological properties of P. massoniana bark and needles for the treatment of rheumatism, intestinal parasites, hypertension, neurasthenia, skin complaints and cancer [11–14].
Pinus species, including P. massoniana, are natural hosts for diverse ectomycorrhizal fungal species [15–19]. It has been reported that ectomycorrhizal symbiosis establishment in the root system confers improved growth performance and tolerance to biotic and abiotic stress in host plants [20–22]. This has been ascribed to the improved nutrient and water acquisition, photosynthetic rate and enhancement of the antioxidant systems and immune system in the hosts [23, 24]. In Pinus, previous studies have shown that ectomycorrhizal fungi improve plant growth and tolerance to drought stress, salinity stress, low-phosphorous stress, heavy metals toxicity, etc. [4, 6, 25–28].
Aluminum (Al) toxicity driven by soil acidification is a long-lasting problem causing forest dieback in many regions of the world [29], particularly in China where natural and forest plantations are declining [30]. Acid rain and anthropogenic soil acidification caused by long-range air pollution and intensive uses of acid-forming fertilizers create nutrient depletion in the soil and accelerate bio-availability of toxic elements such as Al3+ [31–33]. Von Uexkuell and Mutert estimated early in 1995 that acid soils cover more than 70% of potential arable soils [34] but this value could be higher nowadays with the ever-growing industrialization and intensive agriculture. High amount of Al in the soil inhibits plant root growth and decreases nutrient and water uptake [35, 36], leading to significant reductions of plant productivity. It also increases the levels of reactive oxygen species (ROS), leading to lipid peroxidation and cell death [37]. Deciphering the mechanisms of Al tolerance in plants has catalyzed numerous studies and our understanding on the topic is increasing. For example, the exclusion and internal detoxification of Al have been demonstrated in several plants [38–40] and some related genes such as the aluminum activated malate transporter (ALMT) and multidrug and toxic compound extrusion (MATE) have been discovered [40–45].
In forest tree species, some degrees of tolerance to Al toxicity have been observed with a variation among species and genotypes [46–48]. For example, Betula pendula is able to tolerate Al concentration up to 3 mM [49], while Liu and Liu [47] found that the lowest concentration of aluminum toxicity in P. massoniana was 0.15 mM. Besides the intrinsic capacity to exclude Al from root of each plant species, ectomycorrhizal symbiosis establishment in the root system can provide another layer of defensive force against Al stress damages to the host plant. In support to this idea, previous studies have shown that ectomycorrhizal fungi alleviate Al stress damages in Pinus [50, 51]. However, the underlying molecular mechanisms and key genes induced by ectomycorrhizal fungi inoculation under Al stress in Pinus have not been explored.
In a preliminary experiment, we found that inoculation with the ectomycorrhizal fungus species Suillus luteus (SL) promotes P. massoniana growth and imparts Al stress tolerance. We therefore designed the present study to explore the transcriptome of SL inoculated plants and non-inoculated plants under Al stress in order to detect candidate genes mediated by SL conferring Al stress tolerance in P. massoniana.
Result
Morpho-biochemical responses to aluminum stress with or without ectomycorrhizal fungus inoculation
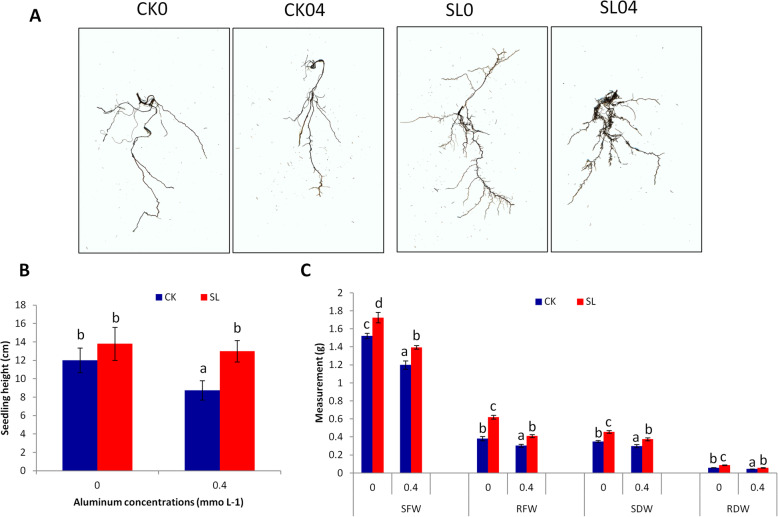
P. massoniana seedlings were subjected to Aluminum (Al) stress for 60 days with Suillus luteus (SL) inoculation or without inoculation (CK). Several morpho-biochemical parameters were investigated. As shown in Fig. 1a, the root density of the seedlings inoculated with the ectomycorrhizal fungus (SL) was higher as compared to non-inoculated seedlings (CK) independently of the Al concentrations, demonstrating the successful inoculation in our experiment. Quantitative analysis of root traits showed that Al treatment reduced all root traits. However, SL inoculation improved the root surface area, average root diameter and root number traits under Al treatment as compared to CK plants (Table 1). Without Al, the seedling height was slightly increased in SL inoculated plants as compared to CK but not significantly (Fig. 1b). Other traits such as shoot fresh weight (SFW), root fresh weight (RFW), shoot dry weight (SDW), root dry weight (RDW) were significantly improved by SL inoculation (Fig. 1c). With 0.4 mmol L− 1 Al application, all morphological traits were reduced independently of SL inoculation (Fig. 1b, c), indicating that Al affects P. massoniana seedling growth. Noteworthy, all morphological traits were significantly higher in SL inoculated plants than in CK plants, implying that SL improves P. massoniana tolerance to Al stress (Fig. 1b, c).
Fig. 1.
Morphological parameters of Pinus massoniana inoculated with Suillus luteus (SL) and non-inoculated plants (CK) under Al stress. a Scans of the root samples from SL inoculated plants and CK plants under 0 and 0.4 mmol L− 1 Al, b seedling height, c shoot fresh weight (SFW), root fresh weight (RFW), shoot dry weight (SDW), root dry weight (RDW). Bars represent average values ± SD of 10 replicates. Different letters above bars indicate significant difference at P < 0.05
Table 1.
Characterization of root traits in Pinus seedlings. Different letters display significant difference at P < 0.05
| Samples | Main root length (cm) | Root surface area (cm2) | Average root diameter (mm) | Root number |
|---|---|---|---|---|
| CK0 | 6.24 ± 1.41a | 0.73 ± 0.8a | 0.36 ± 0.10b | 50.75 ± 8.04b |
| CK04 | 3.98 ± 0.80b | 0.39 ± 0.12c | 0.20 ± 0.05c | 30.00 ± 8.11c |
| SL0 | 5.41 ± 1.30a | 0.77 ± 0.08a | 0.55 ± 0.07a | 63.25 ± 11.45a |
| SL04 | 3.64 ± 0.70b | 0.52 ± 0.15b | 0.39 ± 0.05b | 47.00 ± 9.90b |
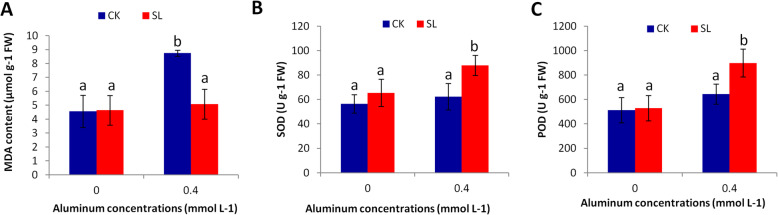
Under stress conditions, plants accumulate excessive levels of reactive oxygen species (ROS). Malondialdehyde (MDA) content is associated with lipid peroxidation via an increased generation of ROS [52]. A high level of MDA is an indicator of a high level of stress damage. We measured the MDA content in needle samples from the four treatments (Fig. 2a). Without Al stress, similar levels of MDA could be observed in CK and SL inoculated plants. However under Al stress, although MDA content increased in both CK and SL inoculated plants, a significantly higher MDA content was measured in CK plants compared to SL inoculated plants. This result suggests that CK plants suffered from oxidative damages under Al stress while SL inoculation helped to keep MDA level in a normal range.
Fig. 2.
Enzyme activities and malondialdehyde (MDA) content in Pinus massoniana inoculated with Suillus luteus (SL) and non-inoculated plants (CK) under Al stress. a MDA content, b Superoxide dismutase (SOD) activity, c, peroxidase (POD) activity in plants grown under 0 and 0.4 mmol L− 1 Al. Bars represent average values ± SD of 10 replicates. Different letters above bars indicate significant difference at P < 0.05
To effectively combat ROS excessive accumulation in plants, a strong activation of antioxidant enzymes such as superoxide dismutase (SOD) and peroxidase (POD) is essential [53]. In this study, we observed that both CK and SL inoculated plants increased their SOD and POD activities under Al stress. However, significantly higher SOD and POD activities were noticed in SL inoculated plants as compared to CK plants (Fig. 2b, c). Altogether, our results suggest that SL inoculation enhances Al stress tolerance in P. massoniana through enhanced antioxidant enzymes activity.
Portray of the transcriptome sequencing and assembly
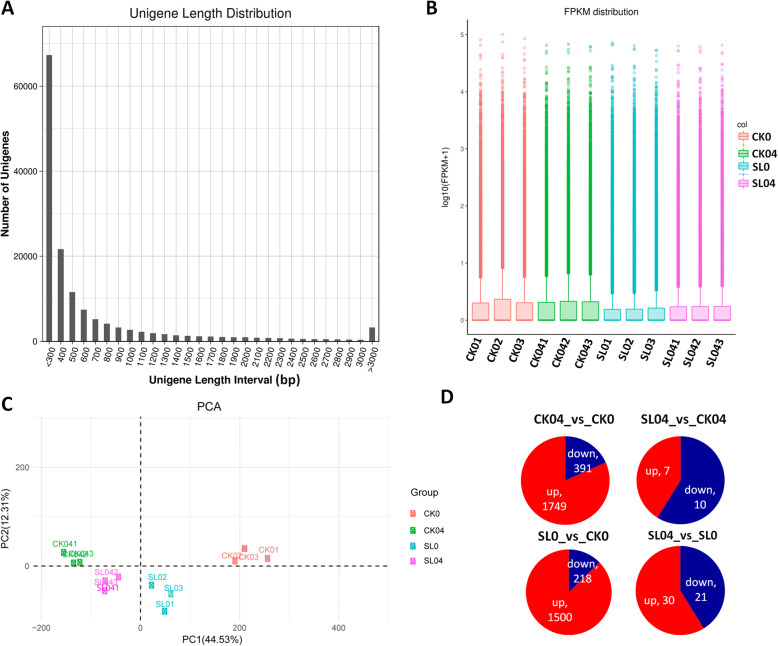
In order to get insight into the molecular basis of Al stress tolerance induced by SL inoculation in P. massoniana, we sequenced the transcriptome of needle samples from the four treatments. With three replicates in each treatment, 12 samples in total were sequenced, yielding on average 54,226,624 raw reads per sample (Table 2). After cleaning, we obtained 89 Gb data with Q30 quality score higher than 93% and error rate lower than 0.03. The clean data were de novo assembled as the reference gene set using the Trinity software and 145,434 unigenes spanning 90,173,863 bp long were obtained with a mean length of 620 bp and a N50 of 1022 bp long. The unigene length distribution is shown in Fig. 3a. The unigenes were annotated in five different databases including NR, SwissProt, PFAM, GO and KO, with 67% of the total unigenes annotated in at least one database (Table 3).
Table 2.
Statistics of the transcriptome sequencing and quality check
| Samples | Raw_reads | Clean_reads | Clean_bases (Gb) | Error (%) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| CK01 | 50,686,474 | 50,295,380 | 6.99 | 0.02 | 97.87 | 93.76 | 47.07 |
| CK02 | 50,394,888 | 50,015,274 | 6.94 | 0.02 | 97.95 | 93.92 | 47.14 |
| CK03 | 61,303,972 | 60,958,750 | 8.43 | 0.029 | 97.99 | 94.03 | 47.34 |
| CK041 | 50,440,018 | 50,157,384 | 6.86 | 0.029 | 97.96 | 94 | 47.7 |
| CK042 | 57,788,340 | 57,453,616 | 7.9 | 0.029 | 97.87 | 93.75 | 47.58 |
| CK043 | 58,747,552 | 58,404,162 | 8.02 | 0.029 | 97.87 | 93.76 | 47.68 |
| SL01 | 57,381,552 | 57,042,586 | 7.8 | 0.029 | 97.99 | 94.08 | 47.95 |
| SL02 | 63,618,448 | 63,275,858 | 8.73 | 0.029 | 97.95 | 93.96 | 47.37 |
| SL03 | 59,962,230 | 59,628,096 | 8.24 | 0.029 | 97.96 | 93.93 | 47.35 |
| SL041 | 43,256,016 | 42,968,462 | 5.96 | 0.029 | 97.84 | 93.67 | 46.98 |
| SL042 | 46,580,296 | 46,369,104 | 6.43 | 0.02 | 98.15 | 94.34 | 47.03 |
| SL043 | 50,559,704 | 50,299,706 | 6.98 | 0.02 | 98.06 | 94.19 | 47.03 |
Fig. 3.
Overview of the transcriptome sequencing and quality. a Unigene length distribution, b Boxplots displaying overall distribution of gene expression among samples, c Principle component analysis of expressed genes, d Pie chart showing the number of up- and down-regulated genes in each compared group. SL0, SL04, CK0 and CK04 represent the Suillus luteus (SL) inoculated plants without Al stress application, SL inoculated plants with 0.4 mmol L− 1 Al stress application, plants non-inoculated without Al stress application and plants non-inoculated with 0.4 mmol L− 1 Al stress application, respectively
Table 3.
Statistics of the unigene annotation
| Database | Number of unigenes | Percentage (%) |
|---|---|---|
| Annotated in NR | 56,449 | 38.81 |
| Annotated in SwissProt | 35,420 | 24.35 |
| Annotated in PFAM | 28,256 | 19.43 |
| Annotated in GO | 26,522 | 18.24 |
| Annotated in KEGG | 11,912 | 8.19 |
| Annotated in all Databases | 137 | 0.09 |
| Annotated in at least one Database | 97,440 | 67 |
| Total Unigenes | 145,434 | 100 |
The gene expression was estimated with fragments per kilobase of exon per million fragments mapped (FPKM) and FPKM value > 1 was used as a threshold to determine the expressed genes (Fig. 3b). Using the FPKM data, we performed a principal component analysis (PCA) to check the clustering pattern of the samples from the four treatments and their replicates. As shown in Fig. 3c, PC1 and PC2 together contributed to over 66% of the global variation. PC1 clearly separated Al treated samples and the non treated samples. By PC2, we could observe a separation between SL inoculated samples from the non-inoculated samples. In addition, the replicates of each treatment were found closely clustered, showing that the quality of the transcriptome sequencing was good enough to proceed to further analyses.
Differentially expressed genes
In order to identify the differentially expressed genes (DEG), we cross-compared the gene count between treatments using following screening criteria: |log2 fold change| ≥ 2 [54], and false discovery rate (FDR) correction set at P < 0.05. In total, 2140, 1718, 17 and 51 DEGs were detected in CK04_vs_CK0, SL0_vs_CK0, SL04_vs_CK04 and SL04_vs_SL0, respectively (Fig. 3d). Gene ontology enrichment analysis of the DEGs showed various biological pathways (metabolic process, photosynthesis, oxido-reduction process, nucleotide binding, etc.) affected by Al and SL treatments (Figure S1). Kyoto Encyclopedia of Genes and Genomes enrichment analyses of the DEGs showed that phorphyrin and chlorophyll metabolism, zeatin biosynthesis were the most enriched pathways affected by Al and SL treatments (Figure S2). The comparison CK04_vs_CK0 provides DEGs involved in Al stress response without SL inoculation, while SL04_vs_SL0 provides DEGs involved in Al stress response with SL inoculation. The conspicuous difference in the number of DEGs between these two comparisons (2140 vs 51) indicates that fewer genes were transcriptionally altered by Al under SL inoculation as compared to conditions without SL inoculation. This confirms that SL inoculation confers Al stress tolerance in P. massoniana.
Core conserved genes regulated by Al stress independently of SL inoculation
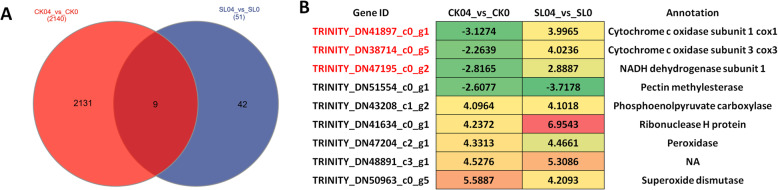
We compared the DEGs induced by Al without SL inoculation (CK04_vs_CK0) and with SL inoculation (SL04_vs_SL0) in order to identify the core regulated DEGs altered by Al stress. As shown in Fig. 4a, nine core DEGs were detected of which, six DEGs (TRINITY_DN51554_c0_g1, TRINITY_DN43208_c1_g2, TRINITY_DN41634_c0_g1, TRINITY_DN47204_c2_g1, TRINITY_DN48891_c3_g1, TRINITY_DN50963_c0_g5) displayed similar patterns of regulation between CK04_vs_CK0 and SL04_vs_SL0 (Fig. 4b), indicating that these genes are essential for Al response in P. massoniana. The gene TRINITY_DN48891_c3_g1 was not functionally annotated, implying it may be an Al-responsive gene specific to P. massoniana. In contrast, we identified three other genes (TRINITY_DN41897_c0_g1 (cox1), TRINITY_DN38714_c0_g5 (cox3) and TRINITY_DN47195_c0_g2 (Nd1)), which displayed opposite patterns of regulation between CK04_vs_CK0 and SL04_vs_SL0 (Fig. 4b). The strong up-regulation of these genes in SL04_vs_SL0 may indicate a tolerance mechanism, which the control plants failed to trigger under Al stress (CK04_vs_CK0).
Fig. 4.
Identification of candidate genes imparting Al stress tolerance in Suillus luteus (SL) inoculated Pinus massoniana seedlings. a Venn diagram depicting the number of shared and specific differentially expressed genes (DEGs) induced by Al without SL inoculation (CK04_vs_CK0) and with SL inoculation (SL04_vs_SL0). b Heatmap showing the log2 fold change of nine core DEGs between CK04_vs_CK0 and SL04_vs_SL0. Gene names highlighted in red are those with an opposite pattern of regulation between the two groups. SL0, SL04, CK0 and CK04 represent the SL inoculated plants without Al stress application, SL inoculated plants with 0.4 mmol L− 1 Al stress application, plants non-inoculated without Al stress application and plants non-inoculated with 0.4 mmol L− 1 Al stress application, respectively
Candidate genes conferring Al stress tolerance induced by SL inoculation
Since P. massoniana seedlings tolerate well Al stress under SL inoculation, we considered the specifically regulated genes in SL04_vs_SL0 as candidate genes imparting Al stress tolerance. In total, 42 DEGs were identified in SL04_vs_SL0 (Fig. 4a, Table 4) but they were not significantly altered in the non-inoculated plants under Al stress (CK04_vs_CK0). Most of the detected genes are involved in antioxidative response, hormone signaling and more importantly plant pathogen infection responses. This shows that prior to exposure to Al stress, SL inoculated plants have an enhanced immune system and ROS scavenging machinery, which may be instrumental for promptly responding to Al stress. Besides, several unannotated DEGs were detected, representing interesting gene resources for future functional characterization.
Table 4.
Genes specifically regulated in SL inoculated plants in response to Al stress
| Gene ID | Log2 Fold Change SL04/SL0 | Annotation |
|---|---|---|
| TRINITY_DN50947_c2_g1 | −10.045 | pre-mRNA-processing protein 40C-like |
| TRINITY_DN42182_c1_g1 | −6.626 | NA |
| TRINITY_DN41191_c0_g1 | −6.3241 | Pectin methylesterase (PME) |
| TRINITY_DN40212_c0_g1 | −5.8947 | NA |
| TRINITY_DN36359_c1_g1 | −5.8146 | Olee1-like protein |
| TRINITY_DN36491_c0_g1 | −5.6695 | NA |
| TRINITY_DN40533_c0_g1 | −4.9629 | DUF538 |
| TRINITY_DN47888_c0_g1 | −4.792 | DUF3799 |
| TRINITY_DN44356_c0_g1 | −3.6987 | Pathogenesis-related protein |
| TRINITY_DN53237_c4_g1 | −2.957 | Subtilisin-like protease SBT5.6 |
| TRINITY_DN10972_c0_g1 | −2.5905 | NA |
| TRINITY_DN44990_c0_g2 | −2.5865 | Ubiquitin |
| TRINITY_DN49726_c1_g1 | −2.4029 | C2 domain |
| TRINITY_DN49726_c1_g2 | −2.3392 | NA |
| TRINITY_DN44727_c0_g4 | −2.2457 | UDP-rhamnose:rhamnosyltransferase 1 |
| TRINITY_DN44741_c1_g1 | −2.1912 | NA |
| TRINITY_DN50117_c0_g5 | −2.1671 | Downy mildew resistant 6 oxygenase |
| TRINITY_DN47573_c2_g4 | −2.0719 | NA |
| TRINITY_DN46639_c1_g3 | −2.0478 | NA |
| TRINITY_DN50628_c0_g3 | −2.0341 | DUF674 |
| TRINITY_DN47580_c3_g1 | 2.0214 | Protein phosphatase 2C |
| TRINITY_DN40477_c0_g2 | 2.1501 | Superoxide dismutase (SOD) |
| TRINITY_DN52128_c2_g4 | 2.2455 | Chaperone protein ClpB1-like protein |
| TRINITY_DN48901_c0_g1 | 2.3314 | 60S acidic ribosomal protein P0 |
| TRINITY_DN46679_c1_g3 | 2.3589 | NA |
| TRINITY_DN49138_c0_g1 | 2.5076 | Plant protein 1589 of unknown function (A_thal_3526) |
| TRINITY_DN45082_c2_g2 | 2.6635 | NA |
| TRINITY_DN47946_c1_g6 | 3.0243 | NA |
| TRINITY_DN50109_c1_g2 | 3.131 | NA |
| TRINITY_DN51142_c2_g1 | 3.2377 | Serine carboxypeptidase-like 40 |
| TRINITY_DN48446_c0_g1 | 3.299 | Peroxidase (POD) |
| TRINITY_DN51077_c0_g4 | 3.3457 | LRR receptor-like serine/threonine-protein kinase |
| TRINITY_DN38876_c0_g1 | 3.4286 | NA |
| TRINITY_DN50590_c0_g1 | 3.5695 | Superoxide dismutase (SOD) |
| TRINITY_DN49159_c3_g1 | 4.3167 | Indole-3-acetic acid (IAA) |
| TRINITY_DN47375_c2_g1 | 4.7598 | NBS-LRR protein G6229 |
| TRINITY_DN49087_c1_g2 | 4.8788 | TIR-NBS-LRR protein |
| TRINITY_DN39211_c0_g1 | 7.2334 | Multidrug and toxic compound extrusion (MATE) |
| TRINITY_DN45197_c0_g1 | 7.3014 | Gluthatione S transferase (GST) |
| TRINITY_DN47067_c5_g1 | 8.1155 | Peroxidase (POD) |
| TRINITY_DN49617_c1_g1 | 10.6606 | Chaperone protein ClpB1 |
| TRINITY_DN44453_c0_g2 | 21.5957 | NA |
qRT-PCR validation of selected genes
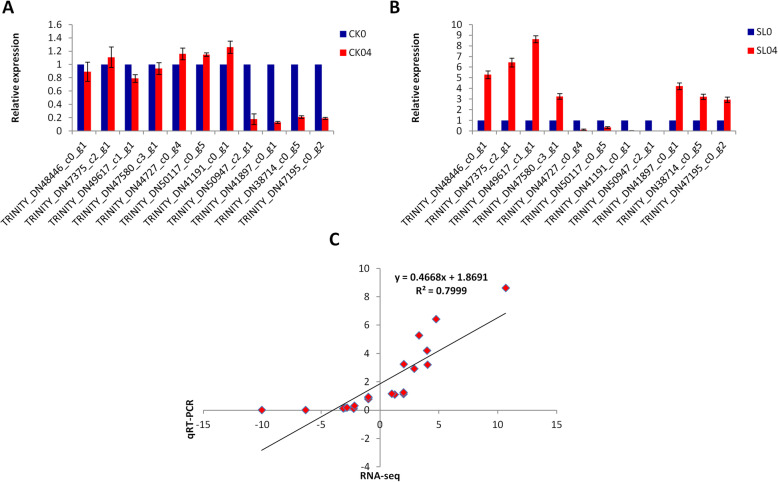
We selected 11 various candidate genes involved in Al stress response in P. massoniana to validate their expression levels using qRT-PCR. The gene Actin2 was used as internal control for expression normalization. The qRT-PCR result showed that the expression levels of all selected genes were significantly altered by Al stress (Fig. 5). In addition, qRT-PCR results were strongly correlated with the RNA-seq report (R2 = 0.8), indicating that the interpretation of the RNA-seq report in this study is reliable.
Fig. 5.
qRT-PCR validation of selected candidate genes. a non-inoculated plants, b inoculated plants. The x-axis represents the genes while the y-axis represents the relative expression of each gene. The bars show standard deviation. c Pearson correlation between qRT-PCR and RNA-seq data. SL0, SL04, CK0 and CK04 represent the Suillus luteus (SL) inoculated plants without Al stress application, SL inoculated plants with 0.4 mmol L− 1 Al stress application, plants non-inoculated without Al stress application and plants non-inoculated with 0.4 mmol L− 1 Al stress application, respectively
Discussion
Suillus luteus inoculation confers aluminum stress tolerance in P. massoniana seedlings
By applying Aluminum (Al) concentrations higher than 0.1 mM, Liu and Liu [47] found that P. massoniana growth is significantly reduced. In this study, we observed a significant inhibition of P. massoniana growth under 0.4 mM Al (Fig. 1; Table 1). Since high Al concentrations (> 1 mM) were reported in forest soils in southern China [30, 55], it is evident that P. massoniana suffers from Al toxicity in the field. Various ectomycorrhizal fungi have been shown to improve Pinus growth performance and tolerance to diverse biotic and abiotic stresses, including Al toxicity [4, 6, 25–28]. Suillus luteus (SL) is an ectomycorrhizal fungus widely found in Pinus plantations [56, 57]. It has been reported that SL can grow in adverse soil conditions such as soils with high salinity, water scarcity, high Mg, Zn, Cd, Pb, Ni, Al [6, 50, 51, 56, 58, 59]. In particular, numerous studies have demonstrated that SL is an Al tolerant fungal species [60–63] and it can facilitate the regeneration and plantation of Pinus seedlings in Al contaminated areas. The results of this study fully corroborate previous findings because our SL inoculated P. massoniana plants tolerated well Al stress (Fig. 1). Yamamoto et al. [37] revealed that high amount of Al induces excessive reactive oxygen species (ROS) accumulation in cells, which ultimately leads to cell death. The weak malondialdehyde content coupled with strong antioxidant enzymes activity in SL inoculated plants (Fig. 2) suggests that SL limits ROS related damages in P. massoniana. This was further confirmed by the significantly lower number of DEGs found in SL inoculated plants as compared to non-inoculated plants under Al stress (Fig. 3d) [64, 65]. In future studies, it will be important to clarify the physiological mechanisms of Al tolerance induced by SL inoculation in P. massoniana. For example, by determining Al content in various plant tissues before and after Al treatment with and without SL inoculation, we will be able to understand how SL affects Al uptake by P. massoniana.
New gene resources for improving Al tolerance in P. massoniana
The mechanisms of Al response including, reduced Al uptake and detoxification of absorbed Al in the symplasm have been well studied in plants [66]. Several related genes with various biological functions such as stress response, membrane transporter, organic acid metabolism, cell wall modification, signaling, hormones, transcription factors have been identified [45, 66–69]. In this study, we identified nine core genes playing diverse functions in response to Al stress in P. massoniana independently of SL inoculation (Fig. 4). The up-regulation of the genes TRINITY_DN47204_c2_g1 (SOD) and TRINITY_DN50963_c0_g5 (POD) showed that activation of antioxidant enzymes is a basic response to Al stress in P. massoniana. The secretion of organic acids such as citrate, malate and oxalate from the roots detoxifies soil Al [70]. The strong up-regulation of the gene TRINITY_DN43208_c1_g2 (phosphoenolpyruvate carboxylase) which is involved in oxalate metabolism suggests that this mechanism is conserved in P. massoniana as demonstrated in other plant species [71, 72]. We noticed the down-regulation of a pectin methylesterase gene (PME, TRINITY_DN51554_c0_g1) which is known to be involved in cell wall structure and high level of PME correlates with high levels of Al adsorption. Therefore, P. massoniana tends to reduce Al adsorption by repressing the expression of the PME [72, 73]. Three core Al responsive genes (Cytochrome c oxidase (cox)1, cox3 and NADH dehydrogenase subunit 1 (Nad1)) associated with mitochondrial activity displayed opposite patterns between SL inoculated and non-inoculated plants. It is well documented that dysfunction of mitochondrial activity seriously impacts on plant fitness [74, 75] and Yamamoto et al. [76] evidenced that Al stress disrupts mitochondrial functions and provokes high accumulation of ROS in tobacco and pea. In this study, all these genes were up-regulated in SL inoculated plants while a strong down-regulation was observed in non-inoculated plants. We deduced that Al stress-induced shut down of mitochondrial activity may have led to high ROS accumulation and subsequently to the growth inhibition observed in non-inoculated plants. We propose these three genes, pending further validation, as marker genes for screening Al tolerant Pinus genotypes.
When engaged in symbiosis with tolerant ectomycorrhizal fungi, plant responses to Al are ameliorated since fungi can immobilize toxic Al into the mycorrhizal roots and surrounding soil environment and improve plant mineral nutrition to ensure normal growth [77]. However, genes mediated by ectomycorrhizal fungi in response to Al stress in host plants have been poorly investigated [23]. In this study, we identified 42 candidate genes unique to SL inoculated plants that may confer Al stress tolerance (Table 4). Most of these genes are involved in Al well known responsive pathways such as antioxidative response, hormone signaling, transporters and plant pathogen infection responses [66]. For example, peroxidase, gluthatione S transferase, multidrug and toxic compound extrusion, indole-3-acetic acid, superoxide dismutase, protein phosphatase 2C, pectin methylesterase have been reported to regulate Al response in plants [45, 54, 72, 73, 78, 79]. A major finding in this study was the numerous genes involved in defense and plant pathogen infection pathway (Table 4). Similar to our results, Luo et al. [23] reported that ectomycorrhizal fungi induced high number of defensive and pathogen infection metabolites and genes under salinity stress in poplar. They concluded that ectomycorrhizal fungi boost the host immune system by priming roots for increased salt tolerance. We speculate that a similar mechanism was established in SL inoculated P. massoniana plants in this study and it helped seedlings to promptly and efficiently combat Al stress effects.
Conclusions
Inoculation of P. massoniana with the ectomycorrhizal fungus Suillus luteus (SL) improves seedling growth and confers Aluminum (Al) stress tolerance. Disruption of the mitochondrial functions through down-regulation of key genes such as cox1, cox2 and Nad1 is probably critical for Al inhibition of growth in non SL inoculated seedlings. Therefore, strategies to activate these genes under Al stress in P. massoniana should be further investigated. We also identified several candidate genes including some unannotated genes that may play cardinal roles in Al stress tolerance. Functional characterization of these gene resources will provide necessary tools for engineering Al tolerant Pinus plants with less dependence on ectomycorrhizal fungi.
Method
Plant material, ectomycorrhizal fungus and stress treatment
P. massoniana Lamb. was used as plant material in this study. The seeds were collected in November 2018 from a superior provenance tree (20 years) planted in Duyun City, Guizhou Province, China. The ectomycorrhizal fungus species Suillus luteus (SL), identified as an aggressive colonizer to P. massoniana [56], was used for plant inoculation. The fruiting part of the fungus was collected from was collected from P. massoniana pure plantation in Longli Forest Farm (N26°28′01″, E107°00′37″), Longli County, Guiyang City, Guizhou Province, China. The formal identification of the plant material and fungus was undertaken by the corresponding author of this article (Professor Guijie Ding). Plant material is available at the National base of P. massoniana (N26°169′ ~ 26°170′, E107°623′ ~ 107°624′) at Maanshan forest farm, Duyun City, Guiyang City, Guizhou Province and a voucher specimen has been deposited at Guizhou Botanical Garden, Guiyang, China, under the accession number: xgnk-2003-a12. No permission was necessary to collect such samples. Inoculum preparation and plant inoculation were performed following detailed descriptions in works of Yu et al. [6]. Seedlings were raised from February to July 2019 and 6-months old seedlings inoculated with S. luteus (inspected for successful mycorrhization) were used as test materials while 6-months old seedlings without inoculation were used as control.
The experiment was carried out in a greenhouse with light intensity of 600–800 μmol m− 2 s− 1, relative humidity of 55%, photoperiod of 16 h, 25 °C at light and 18 °C in the dark. The quartz sand was rinsed, sterilized in an autoclave (pressure 0.14 MPa, 121 °C) for 2 h, and then loaded into plastic pots (21 cm × 15 cm × 18.5 cm). Uniform seedlings were selected and transplanted into the pots. They were kept under normal growth conditions for 2 weeks by pouring frequently 1/2 Hoagland nutrient solution. Then, the stress treatment started. Four treatments were set, namely 2 inoculated treatments SL0 (0 mmol L− 1 Al3+), SL04 (0.4 mmol L− 1 Al3+) and 2 non-inoculated treatments CK0 (0 mmol L− 1 Al3+), CK04 (0.4 mmol L− 1 Al3+) following descriptions of Liu and Liu [47]. For Aluminum (Al) stress, Al was added to total Hoagland nutrient solution in the form of anhydrous AlCl3 and the pH value was adjusted to 4.1 ± 0.1 [47] with 0.1 M diluted HCl or NaOH to maintain acidic conditions. In order to maintain the Al activity, 0.5 mmol L− 1 CaCl2 was added into the nutrient solution at the same time to avoid interaction between Al ion and solution ions. The treatment solution was poured once a week. Each treatment was repeated five times containing 15 pots each, with three plants per pot. Plants were collected on the 60th day after the induction of Al stress to measure the growth and physiological parameters in the four groups. In addition, we randomly selected three plants from different replicates in each treatment and needle samples were collected for transcriptome analysis. Samples were frozen in liquid nitrogen and stored at − 80 °C for further use.
Measurement of morphological parameters
Ten plants per treatment were selected from the five replicates (two plants per replicate), gently shaken to remove the quartz sand on the root surface, and carefully washed with running water. Root quantitative traits were measured from scanned images from a desktop scanner (EPSON Perfection V800 Photo, CA, USA) using the WinRHIZO Pro software (Regent Instruments Inc., Quebec, Canada). Seedling height was measured with a vernier caliper; the root and shoot fresh weights were recorded separately using an electric balance (Type: ML 204; Mettler Toledo Company, Greifensee, Switzerland; measurement accuracy 0.0001 g). Samples were dried at 80 °C to constant weight and the dry weights were recorded.
Determination of SOD, POD, soluble sugar and MDA
The content of Malondialdehyde (MDA) and the activities of superoxide dismutase (SOD) and peroxidase (POD) in the needle samples from the four treatments were determined in five replicates separately according to the instructions provided by kits (COMIN, Keming Biotechnology Co. Ltd., Suzhou, China). The content of MDA was expressed as μmol g− 1 FW. SOD and POD activities were expressed as units per gram fresh weigh (U g− 1 FW). One unit (U) of SOD activity was defined as the activity of SOD when the inhibition of the xanthine oxidase coupling reaction system was 50%, while one unit (U) of POD activity was defined as the variation of 0.01 in A470 per mL reaction system.
Statistical analysis
All data were statistically analyzed with Minitab18 software (P < 0. 05). Tukey’s test based on analysis of variance (ANOVA) was chosen to analyze the data.
RNA extraction, cDNA library construction and transcriptome sequencing
Experiments are conducted following standard procedures of Shanghai Applied Protein Technology, Co., Ltd. (APT, Shanghai, China). Briefly, total RNAs were extracted from 12 needle samples with RNAprep Pure Plant Kit (TIANGEN, Beijing, China). Concentrations of the samples were quantified by a NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific). To get high quality RNA, samples were tested on 1% agarose gel electrophoresis for the integrity of RNA and DNA contamination. For accurate detection of RNA integrity, Agilent 2100 Bioanalyzer was used. RNA quantification was performed using Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, Carlsbad, CA, USA). Next, RNA integrity was checked by the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). For cDNA synthesis, a total 1 μg RNA for each sample was treated with DNase I to eradicate the genomic DNA and then used as a template for reverse transcription (QuantiTect Reverse Transcription Kit, Qiagen, China).
We added fragment buffer to break into short segments using short segment RNA as template. Sequencing libraries was created using NEB Next Ultra RNA Library Prep Kit following manufacturer′s instructions. The index codes were added to each sample. Briefly, the mRNA was purified from 3 μg total RNA of each of three replicate using poly-T oligo-attached magnetic beads and then broken into short fragments to synthesize first strand cDNA. The second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. PCR was carried out with Phusion High Fidelity DNA polymerase using universal PCR primers and index (×) primer. Finally, 12 paired-end cDNA libraries with an insert size of 300 bp were constructed for transcriptome sequencing and sequenced on Illumina HiSeq 4000 platform (Illumina Inc., San Diego, USA) at Shanghai Applied Protein Technology, Co., Ltd. (APT, Shanghai, China).
De novo assembly, functional annotation, expression analysis
The clean reads were obtained after trimming adapter sequences, removal of low quality (containing > 50% bases with a Phred quality score < 20) and reads with unknown nucleotides (more than 1% ambiguous residues N) using the FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Analysis of the GC content distribution was performed. Transcriptome assembly was performed using Trinity r20140717 [80] and employing paired-end method. For hierarchical clustering, Corset was used (https://code.google.com/p/corset-project/). The longest cluster sequence was obtained by clustering with Corset hierarchy as unigene for subsequent analysis. The assembled unigenes were then annotated in various databases such as KEGG using KAAS (E-value < 1.0 × 10− 10), GO using Blast2GOv2.5 (E-value < 1.0 × 10− 6), PFAM using HMMER3.0 (E-value < 0.01), Swissprot using BLAST 2.6.0+ (E-value < 1.0 × 10− 5) and NR using BLAST 2.6.0+ (E-value < 1.0 × 10− 5).
The sequenced reads were compared with the unigene library using Bowtie2 [81], and the level of expression was estimated in combination with RSEM [82]. The gene expression level was determined according to the fragments per kilobase of exon per million fragments mapped (FPKM) method. The Principal component analysis was performed in R v2.3.0. The read count was normalized and EdgeR Bioconductor package [83], was used to determine the differential expression genes (DEGs) between groups with the |log2 fold change| ≥ 2 and false discovery rate (FDR) correction set at P < 0.05 [54]. GO enrichment analysis was performed using the topGO method based on the wallenius noncentral hypergeometric distribution with P < 0.05 [84]. KEGG pathway enrichment analysis of the DEGs was done using KOBAS2.0 [85]. The FDR correction was employed (P < 0.05) to reduce false positive prediction of enriched GO terms and KEGG pathways.
Gene expression analysis using quantitative real PCR
In order to confirm the gene expression levels obtained from the RNA-seq, a qRT-PCR analysis was performed on RNA extracted from needle samples as described previously [86]. The qRT-PCR was conducted on a Roche Lightcyler® 480 instrument using the SYBR Green Master Mix (Vazyme, Vazyme Biotech Co. Ltd., Nanjing, China) following the manufacturer’s protocol. The gene Actin2 was used as internal control. Specific primer sequences for selected genes were designed with PrimerPremier 5 and are presented in Table S1.
Supplementary Information
Additional file 1: Table S1. Primer sequences used for qRT-PCR in this study.
Additional file 2: Figure S1. GO enrichment analysis. SL0, CK0 and CK04 represent the Suillus luteus (SL) inoculated plants without Al stress application, SL inoculated plants with 0.4 mmol L− 1 Al stress application, plants non-inoculated without Al stress application and plants non-inoculated with 0.4 mmol L− 1 Al stress application, respectively. Figure S2. KEGG enrichment analysis. SL0, CK0 and CK04 represent the Suillus luteus (SL) inoculated plants without Al stress application, plants non-inoculated without Al stress application and plants non-inoculated with 0.4 mmol L− 1 Al stress application, respectively.
Acknowledgements
Not applicable.
Abbreviations
- Al
Aluminum
- SL
Suillus luteus
- ROS
Reactive oxygen species
- MDA
Malondialdehyde
- SOD
Superoxide dismutase
- POD
Peroxidase
- FPKM
Fragments per kilobase of exon per million fragments mapped
- PCA
Principal component analysis
- DEG
Differentially expressed genes
- FDR
False discovery rate
Authors’ contributions
HL: experimental set up, RNA-seq data analysis and manuscript writing; HC and KL: physiological and biochemical indicators analysis; GD: qRT-PCR validation, RNA-seq data analysis manuscript writing; QR: Study conception, funding acquisition, supervision, revision of the manuscript. All authors have read and approved the manuscript.
Funding
This work was funded by the National Key Research and Development Program of China (No. 2017YFD06003002) and the Science and Technology Foundation of Guizhou Province ([2016]1144). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the NCBI Bioproject repository, accession number: PRJNA636599. The data will be released upon publication of this manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Luo YH, Sun DJ, Lin JY, Guo WF, Lu LH, Wen YG. Effect of close-to-nature management on the natural regeneration and species diversity in a masson pine plantation. Acta Ecol Sin. 2013;33(19):6154–6162. doi: 10.5846/stxb201306101601. [DOI] [Google Scholar]
- 2.Dou X, Deng Q, Li M, Wang W, Zhang Q, Cheng X. Reforestation of Pinus massoniana alters soil organic carbon and nitrogen dynamics in eroded soil in South China. Ecol Eng. 2013;52:154–160. doi: 10.1016/j.ecoleng.2012.12.099. [DOI] [Google Scholar]
- 3.Wang Y, Ding G. Physiological responses of mycorrhizal Pinus massoniana seedlings to drought stress and drought resistance evaluation. Chin J Appl Ecol. 2013;24:639–645. [PubMed] [Google Scholar]
- 4.Zhang, T., Wen, X. & Ding, G. Ectomycorrhizal symbiosis enhances tolerance to low phosphorous through expression of phosphate transporter genes in masson pine (Pinus massoniana). Acta Physiol Plant 39, 101 (2017). 10.1007/s11738-017-2392-y.
- 5.Wang X, Lu L, Xing H, Zeng J, Xie Y, Cai D, Liu X, Zhang X. Effects of close-to-nature conversion on Pinus massoniana plantations at different stand developmental stages. Trop Conserv Sci. 2018;11:1–16. doi: 10.1177/1940082918767953. [DOI] [Google Scholar]
- 6.Yu P, Sun Y, Huang Z, Zhua F, Sun Y, Jiang L. The effects of ectomycorrhizal fungi on heavy metals’ transport in Pinus massoniana and bacteria community in rhizosphere soil in mine tailing area. J Hazard Mater. 2020;381:121203. doi: 10.1016/j.jhazmat.2019.121203. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Z, Zhang SY. China's plantation forests for sustainable wood supply and development. 2003. [Google Scholar]
- 8.Yang H, Wang S, Zhang J, Fan B, Zhang W. Biomass and nutrients of Pinus massoniana plantations in southern China: Simulations for different managing practices. J Food Agric Environ. 2011;9:689–693. [Google Scholar]
- 9.Deng X, Zhang L, Lei P, Xiang W, Yan W. Variations of wood basic density with tree age and social classes in the axial direction within Pinus massoniana stems in Southern China. Ann Forest Sci. 2014;71(4):505–516. doi: 10.1007/s13595-013-0356-y.hal-01101774. [DOI] [Google Scholar]
- 10.Zhang, P., He, Y., Feng, Y., De La Torre R., Jia H., Tang J., Cubbage F. An analysis of potential investment returns of planted forests in South China. New For 50, 943–968 (2019). 10.1007/s11056-019-09708-x.
- 11.Cui Y, Xie H, Wang J. Potential biomedical properties of Pinus massoniana bark extract. Phytother Res. 2005;19:34–38. doi: 10.1002/ptr.1619. [DOI] [PubMed] [Google Scholar]
- 12.Ma H, Lai F, Xie H, Wang J, Wang H. Involvement of the Bcl-2 family members in Pinus massoniana bark extract induced apoptosis in HeLa cells. Phytother Res. 2008;22:1472–1476. doi: 10.1002/ptr.2496. [DOI] [PubMed] [Google Scholar]
- 13.Duke JA, Ayensu ES. Medicinal Plants of China. 1985;2(705 S):1300. Strichzeichnungen: Reference Publ., Inc. Algonac. Michigan. ISBN 0‐917266‐20‐4.
- 14.Wu D, Li S, Yang D, Cui Y. Effects of Pinus massoniana bark extract on the adhesion and migration capabilities of HeLa cells. Fitoterapia. 2011;82:1202–1205. doi: 10.1016/j.fitote.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Rincón, A., Álvarez, I. & Pera, J. Ectomycorrhizal fungi of Pinus pinea L. in northeastern Spain. Mycorrhiza 8, 271–276 (1999). 10.1007/s005720050245.
- 16.Rincón A, Alvarez IF, Pera J. Inoculation of containerized Pinus pinea L seedlings with seven ectomycorrhizal fungi. Mycorrhiza. 2001;11:265–271. doi: 10.1007/s005720100127. [DOI] [PubMed] [Google Scholar]
- 17.Parladé J, Pera J, Alvarez IF. Inoculation of containerized Pseudotsuga menziesii and Pinus pinaster seedlings with spores of five species of ectomycorrhizal fungi. Mycorrhiza. 1996;6:237–245. doi: 10.1007/s005720050131. [DOI] [Google Scholar]
- 18.Duñabeitia MK, Hormilla S, Garcia-Plazaola JI, Txarterina K, Arteche U, Becerril JM. Differential responses of three fungal species to environmental factors and their role in the mycorrhization of Pinus radiata. Mycorrhiza. 2004;14:11–18. doi: 10.1007/s00572-003-0270-5. [DOI] [PubMed] [Google Scholar]
- 19.Lu N, Yu M, Cui M, Luo Z, Feng Y, Cao S, Sun Y, Li Y. Effects of different Ectomycorrhizal fungal inoculates on the growth of Pinus tabulaeformis seedlings under greenhouse conditions. Forests. 2016;7:316. doi: 10.3390/f7120316. [DOI] [Google Scholar]
- 20.Garbaye J. The role of ectomycorrhizal symbiosis in the resistance of forests to water stress. Outlook Agr. 2000;29:63–69. doi: 10.5367/000000000101293068. [DOI] [Google Scholar]
- 21.Kohler J, Hernández JA, Caravaca F, Roldán A. Induction of antioxidant enzymes is involved in the greater effectiveness of a PGPR versus AM fungi with respect to increasing the tolerance of lettuce to severe salt stress. Environ Exp Bot. 2009;65:245–252. doi: 10.1016/j.envexpbot.2008.09.008. [DOI] [Google Scholar]
- 22.Lehto T, Zwiazek JJ. Ectomycorrhizas and water relations of trees: a review. Mycorrhiza. 2011;21:71–90. doi: 10.1007/s00572-010-0348-9. [DOI] [PubMed] [Google Scholar]
- 23.Luo Z, Janz D, Jiang X, Göbel C, Wildhagen H, Tan Y, Rennenberg H, Feussner I, Polle A. Upgrading root physiology for stress tolerance by Ectomycorrhizas: insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiol. 2009;151:1902–1917. doi: 10.1104/pp.109.143735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao, Z., Xie, W., & Chen, B. (2019). Arbuscular Mycorrhizal Symbiosis affects plant immunity to viral infection and accumulation. Viruses, 11(6), 534. https://doi.org/10.3390/v11060534. [DOI] [PMC free article] [PubMed]
- 25.Zhu, J., Li, F., Xu, M., Kang H., Wu X. The role of ectomycorrhizal fungi in alleviating pine decline in semiarid sandy soil of northern China: an experimental approach. Ann For Sci 65, 304 (2008). 10.1051/forest:2008007.
- 26.Wang J, Huang Y, Jiang XY. Influence of ectomycorrhizal fungi on absorption and balance of essential elements of Pinus tabulaeformis seedlings in saline soil. Pedosphere. 2011;21(3):400–406. doi: 10.1016/S1002-0160(11)60141-0. [DOI] [Google Scholar]
- 27.Kipfer T, Wohlgemuth T, van der Heijden MGA, Ghazoul J, Egli S (2012) Growth response of drought-stressed Pinus sylvestris seedlings to single- and multi-species inoculation with Ectomycorrhizal Fungi. PLoS One 7(4): e35275. 10.1371/journal.pone.0035275. [DOI] [PMC free article] [PubMed]
- 28.Yin D, Halifu S, Song R, Qi J, Deng X, Den J Effects of an ectomycorrhizal fungus on the growth and physiology of Pinus sylvestris var mongolica seedlings subjected to saline–alkali stress J For Res 2020, 31, 781–788. 10.1007/s11676-019-01007-7.
- 29.Ulrich B (1989). Effects of acidic precipitation on forest ecosystems in Europe. In: Adriano et al. (eds.). Acidic Precipitation, Biological and Ecological Effects. New York: Springer, 189–272.
- 30.Zhang J, Lyu Z, Shao S, Li F, Yang S, Song W, Li W, Li S. Effects of aluminum toxicity induced by acid deposition on pine Forest ecosystem in Longli of Guizhou Province, Southwestern China. Chin Geogra Sci. 2016;26:495–507. doi: 10.1007/s11769-015-0763-0. [DOI] [Google Scholar]
- 31.Ulrich B. Effects of acidic precipitation on forest ecosystem in Europe. In: Adriano DC, Johnson AH, editors. Acidic Precipitation. Berlin: Spring-Verlay; 1989. pp. 189–272. [Google Scholar]
- 32.Rengel Z. Handbook of soil acidity. New York: Marcel Dekker Inc; 2003. p. 496. [Google Scholar]
- 33.Li W, Johnson CE. Relationships among pH, aluminum solubility and aluminum complexation with organic matter in acid forest soils of the northeastern United States. Geoderma. 2016;271:234–242. doi: 10.1016/j.geoderma.2016.02.030. [DOI] [Google Scholar]
- 34.Von Uexkuell HR, Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171:1–15. doi: 10.1007/BF00009558. [DOI] [Google Scholar]
- 35.Ryan PR, DiTomaso JM, Kochian LV. Aluminium toxicity in roots: an investigation of spatial sensitivity and the role of the root cap. J Exp Bot. 1993;44:437–446. doi: 10.1093/jxb/44.2.437. [DOI] [Google Scholar]
- 36.Kochian LV, Pineros MA, Hoekenga OA. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil. 2005;274:175–195. doi: 10.1007/s11104-004-1158-7. [DOI] [Google Scholar]
- 37.Yamamoto Y, Kobayashi Y, Matsumoto H. Lipid peroxidation is an early symptom triggered by aluminium, but not the primary cause of elongation inhibition in pea roots. Plant Physiol. 2001;125:199–208. doi: 10.1104/pp.125.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol. 2004;55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- 39.Ma JF. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol. 2007;264:225–252. doi: 10.1016/S0074-7696(07)64005-4. [DOI] [PubMed] [Google Scholar]
- 40.Brunner, I., & Sperisen, C. (2013). Aluminum exclusion and aluminum tolerance in woody plants. Front Plant Sci, 4, 172. 10.3389/fpls.2013.00172. [DOI] [PMC free article] [PubMed]
- 41.Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, et al. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004;37:645–653. doi: 10.1111/j.1365-313X.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 42.Hoekenga OA, Maron LG, Piñeros MA, Cançado GM, Shaff J, Kobayashi Y, et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, et al. An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 2007;48:1081–1091. doi: 10.1093/pcp/pcm091. [DOI] [PubMed] [Google Scholar]
- 44.Magalhaes JV, Liu J, Guimarães CT, Lana UGP, Alves VMC, Wang YH, et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 2007;39:1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- 45.Bian M, Zhou M, Sun D, Li C. Molecular approaches unravel the mechanism of acid soil tolerance in plants. Crop J. 2013;1:91–104. doi: 10.1016/j.cj.2013.08.002. [DOI] [Google Scholar]
- 46.Schaedle M, Thornton FC, Raynal DJ, Tepper HB. Response of tree seedlings to aluminum. Tree Physiol. 1989;5:337–356. doi: 10.1093/treephys/5.3.337. [DOI] [PubMed] [Google Scholar]
- 47.Liu R, Liu H. Effect of acidity and aluminum on the growth of Pinus massoniana seedlings. Acta Bot Sin. 1995;37(2):154–158. [Google Scholar]
- 48.Grisel, N., Zoller, S., Künzli-Gontarczyk, M., Lampart, T., Münsterkötter, M., Brunner, I., Bovet, L., Métraux, J. P., & Sperisen, C. (2010). Transcriptome responses to aluminum stress in roots of aspen (Populus tremula). BMC Plant Biol, 10, 185. 10.1186/1471-2229-10-185. [DOI] [PMC free article] [PubMed]
- 49.Göransson A, Eldhuset TD. Effects of aluminium on root growth and nutrient uptake of Betula pendula seedlings. Physiol Plant. 1987;69:193–199. doi: 10.1111/j.1399-3054.1987.tb04275.x. [DOI] [Google Scholar]
- 50.Kasuya CM, Muchovej R, Muchovej J. Influence of aluminum on in vitro formation of Pinus caribaea mycorrhizae. Plant Soil. 1990;124:73–77. doi: 10.1007/BF00010933. [DOI] [Google Scholar]
- 51.Ahonen-Jonnarth, U., Göransson, A., Finlay, R. D. (2003). Growth and nutrient uptake of ectomycorrhizal Pinus sylvestris seedlings in a natural substrate treated with elevated Al concentrations. Tree Physiol, 23(3), 157–167. 10.1093/treephys/23.3.157. [DOI] [PubMed]
- 52.Sheoran S, Thakur V, Narwal S, Turan R, Mamrutha HM, Singh V, Tiwari V, Sharma I. Differential activity and expression profile of antioxidant enzymes and physiological changes in wheat (Triticum aestivum L.) under drought. Appl Biochem Biotechnol. 2015;177(6):1282–1298. doi: 10.1007/s12010-015-1813-x. [DOI] [PubMed] [Google Scholar]
- 53.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17(7):1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H, Lu C, Jiang H, Peng J. Global transcriptome analysis reveals distinct aluminum-tolerance pathways in the Al-accumulating species Hydrangea macrophylla and marker identification. PLoS One. 2015;10(12):e0144927. doi: 10.1371/journal.pone.0144927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo J, David Vogt R, Zhang X, Zhang Y, Seip HM, Xiao J, Tang H. Aluminium Mobilization from Acidic Forest Soils in Leigongshan Area, Southwestern China: Laboratory and Field Study. Arch Environ Contam Toxicol. 2006;51:321–328. doi: 10.1007/s00244-003-0151-0. [DOI] [PubMed] [Google Scholar]
- 56.Huang, J., Nara, K., Lian, C., Zong, K., Peng, K., Xue, S., & Shen, Z. (2012). Ectomycorrhizal fungal communities associated with Masson pine (Pinus massoniana Lamb.) in Pb-Zn mine sites of central South China. Mycorrhiza 22(8), 589–602. 10.1007/s00572-012-0436-0. [DOI] [PubMed]
- 57.Hayward, J., Horton, T. R., Pauchard, A., & Nuñnez, M. A. (2015). A single ectomycorrhizal fungal species can enable a Pinus invasion. Ecology, 96(5), 1438–1444. 10.1890/14-1100.1. [DOI] [PubMed]
- 58.Colpaert JV, Vandenkoornhuyse P, Adriaensen K, Vangronsveld J. Genetic variation and heavy metal tolerance in the ectomycorrhizal basidiomycete Suillus luteus. New Phytol. 2000;147:367–379. doi: 10.1046/j.1469-8137.2000.00694.x. [DOI] [Google Scholar]
- 59.Huang, J., Han, Q., & Li, J. (2018). Soil propagule bank of ectomycorrhizal fungi associated with Masson pine (Pinus massoniana) grown in a manganese mine wasteland. PLoS One, 13(6), e0198628. https://doi.org/10.1371/journal.pone.0198628. [DOI] [PMC free article] [PubMed]
- 60.Thompson GW, Medve RJ. Effects of aluminum and manganese on the growth of ectomycorrhizal fungi. Appl Environ Microbiol. 1984;48(3):556–560. doi: 10.1128/AEM.48.3.556-560.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leski T, Rudawska M, Kieliszewska-Rokicka B. Intraspecific aluminium response in Suillus luteus (L.) s.f. gray., an ectomycorrhizal symbiont of scots pine. Acta Soc Bot Pol. 1995;64:97–105. doi: 10.5586/asbp.1995.014. [DOI] [Google Scholar]
- 62.Li H, Huang JG, Yuan L. Influence of aluminum and manganese on the growth, nutrient uptake and the efflux by Ectomycorrhizal Fungi. Environ Sci. 2013;34(1):315–320. [PubMed] [Google Scholar]
- 63.Wang MX, Yuan L, Huang JG, Zhou ZF. Al3+ Absorption and Assimilation by Four Ectomycorrhizal Fungi. Environ Sci. 2015;36(9):3479–3485. doi: 10.13227/j.hjkx.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 64.Wang, L., Li, D., Zhang, Y., Gao, Y., Yu, J., Wei, X., & Zhang, X. (2016). Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. PLoS One, 11(3), e0149912. 10.1371/journal.pone.0149912. [DOI] [PMC free article] [PubMed]
- 65.Dossa, K., Li, D., Wang, L., Zheng, X., Liu, A., Yu, J., Wei, X., Zhou, R., Fonceka, D., Diouf, D., Liao, B., Cissé, N., & Zhang, X. (2017). Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Sci Rep, 7(1), 8755. 10.1038/s41598-017-09397-6. [DOI] [PMC free article] [PubMed]
- 66.Bojórquez-Quintal, E., Escalante-Magaña, C., Echevarría-Machado, I., & Martínez-Estévez, M. (2017). Aluminum, a friend or foe of higher plants in acid soils. Front Plant Sci, 8, 1767. 10.3389/fpls.2017.01767. [DOI] [PMC free article] [PubMed]
- 67.Guo P, Qi YP, Huang WL, et al. Aluminum-responsive genes revealed by RNA-Seq and related physiological responses in leaves of two Citrus species with contrasting aluminum-tolerance. Ecotoxicol Environ Saf. 2018;158:213–222. doi: 10.1016/j.ecoenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 68.Guo P, Qi YP, Cai YT, Yang TY, Yang LT, Huang ZR, Chen LS. Aluminum effects on photosynthesis, reactive oxygen species and methylglyoxal detoxification in two Citrus species differing in aluminum tolerance. Tree Physiol. 2018;38(10):1548–1565. doi: 10.1093/treephys/tpy035. [DOI] [PubMed] [Google Scholar]
- 69.Guo P, Qi Y-P, Yang L-T, Lai N-W, Ye X, Yang Y, Chen L-S. Root adaptive responses to aluminum-treatment revealed by RNA-Seq in two Citrus species with different aluminum-tolerance. Front Plant Sci. 2017;8:330. doi: 10.3389/fpls.2017.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng SJ, Ma JF, Matsumoto H. High aluminum resistance in buckwheat. I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol. 1998;117:745–751. doi: 10.1104/pp.117.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Begum HH, Osaki M, Watanabe T, Shinano T. Mechanisms of aluminum tolerance in Phosphoenolpyruvate carboxylase transgenic Rice. J Plant Nutr. 2009;32(1):84–96. doi: 10.1080/01904160802531035. [DOI] [Google Scholar]
- 72.Xu L-M, Liu C, Cui B-M, Wang N, Zhao Z, Zhou L-N, Huang K-F, Ding J-Z, Du H-M, Jiang W, Zhang S-Z. Transcriptomic responses to aluminum (Al) stress in maize. J Integr Agric. 2018;17(9):1946–1958. doi: 10.1016/S2095-3119(17)61832-X. [DOI] [Google Scholar]
- 73.Schmohl NJ, Fisahn J, Horst WJ. Pectin methylesterase modulates aluminium sensitivity in Zea mays and Solanum tuberosum. Physiol Plant. 2000;109:419–427. doi: 10.1034/j.1399-3054.2000.100408.x. [DOI] [Google Scholar]
- 74.Mansilla, N., Racca, S., Gras, D. E., Gonzalez, D. H., & Welchen, E. (2018). The complexity of mitochondrial complex IV: an update of cytochrome c oxidase biogenesis in plants. Int J Mol Sci, 19(3), 662. 10.3390/ijms19030662. [DOI] [PMC free article] [PubMed]
- 75.Fromm, S., Senkler, J., Eubel, H., Peterhänsel, C., & Braun, H. P. (2016). Life without complex I: proteome analyses of an Arabidopsis mutant lacking the mitochondrial NADH dehydrogenase complex. J Exp Bot, 67(10), 3079–3093. 10.1093/jxb/erw165. [DOI] [PMC free article] [PubMed]
- 76.Yamamoto Y, Kobayashi Y, Devi SR, Rikiishi S, Matsumoto H. Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 2002;128(1):63–72. doi: 10.1104/pp.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo ZB, Wu CH, Zhang C, Li H, Lipka U, Polle A. The role of ectomycorrhizas in heavy metal stress tolerance of host plants. Environ Exp Bot. 2014;108:47–62. doi: 10.1016/j.envexpbot.2013.10.018. [DOI] [Google Scholar]
- 78.Kumari M, Taylor GJ, Deyholos MK. Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol Gen Genomics. 2008;279:339–357. doi: 10.1007/s00438-007-0316-z. [DOI] [PubMed] [Google Scholar]
- 79.Mattiello L, Kirst M, da Silva FR, Jorge RA, Menossi M. Transcriptional profile of maize roots under acid soil growth. BMC Plant Biol. 2010;10:196. doi: 10.1186/1471-2229-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langmead, B., Salzberg, S. Fast gapped-read alignment with bowtie 2. Nat Methods 9, 357–359 (2012). 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed]
- 82.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1007/978-1-4939-0512-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson, M. D., McCarthy, D. J., & Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed]
- 84.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C-Y, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dossa K, Mmadi MA, Zhou R, Zhang T, Su R, Zhang Y, Wang L, You J, Zhang X. Depicting the core transcriptome modulating multiple abiotic stresses responses in sesame (Sesamum indicum L.). Int. J Mol Sci. 2019;20:2–22. doi: 10.3390/ijms20163930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primer sequences used for qRT-PCR in this study.
Additional file 2: Figure S1. GO enrichment analysis. SL0, CK0 and CK04 represent the Suillus luteus (SL) inoculated plants without Al stress application, SL inoculated plants with 0.4 mmol L− 1 Al stress application, plants non-inoculated without Al stress application and plants non-inoculated with 0.4 mmol L− 1 Al stress application, respectively. Figure S2. KEGG enrichment analysis. SL0, CK0 and CK04 represent the Suillus luteus (SL) inoculated plants without Al stress application, plants non-inoculated without Al stress application and plants non-inoculated with 0.4 mmol L− 1 Al stress application, respectively.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the NCBI Bioproject repository, accession number: PRJNA636599. The data will be released upon publication of this manuscript.