Figure 1.
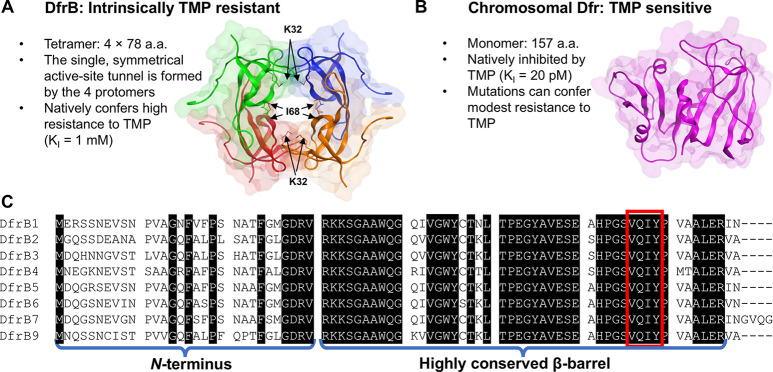
The homotetrameric DfrBs are evolutionarily distinct from the chromosomal Dfrs. (A) The homotetrameric DfrB1 (PDB: 2RK1) is intrinsically TMP-resistant: its large, symmetrical active-site tunnel does not bind TMP. Each protomer is colored differently. The 20 N-terminal residues of each protomer are unstructured and are not represented. For clarity, only I68 of the V66-Q67-I68-Y69 (VQIY) region and K32 are represented as sticks on each protomer. (B) The chromosomal Dfrs are evolutionarily and structurally unrelated. The E. coli Dfr (PDB: 1DDR), shown at scale, is strongly inhibited by TMP. (C) Sequence alignment of the DfrB family; there is no DfrB8.15 The weakly conserved N-termini and the highly conserved β-barrel core including the VQIY residues that line the active site tunnel 4-fold are identified.