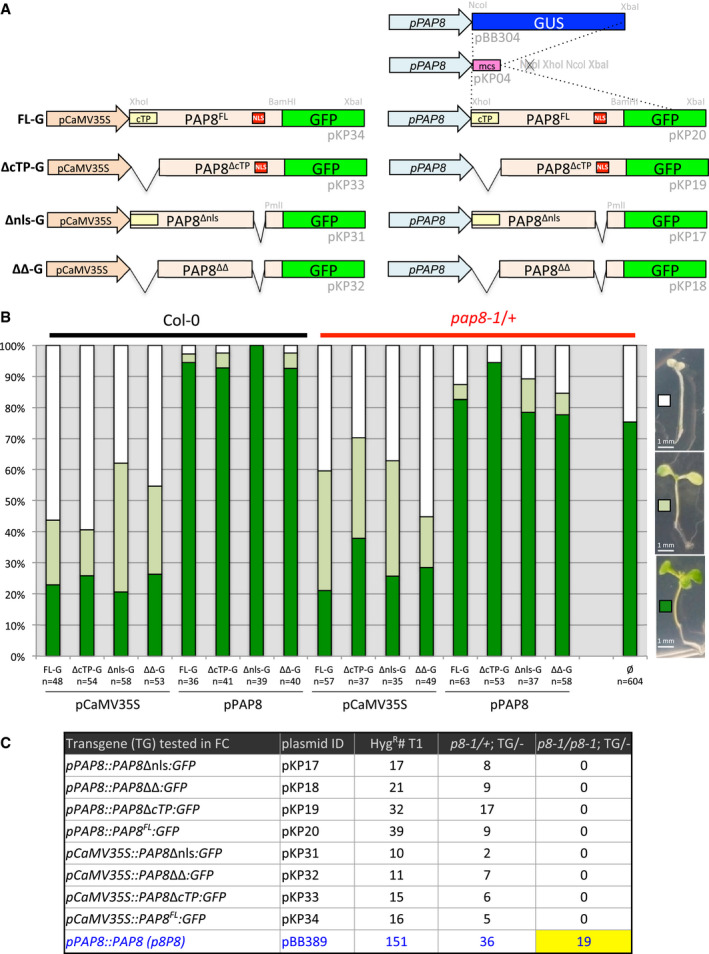
Figure EV3. (Related to Fig 3) PAP8 and its variants fused to GFP .
-
ASchematic illustration of the GFP‐fused PAP8 variants used for sub‐cellular localization and functional complementation tests FL‐G, PAP8 full‐length ORF translationally fused to GFP; Δnls‐G, deletion of the NLS (nuclear localization signal), ORF fused to GFP; ΔcTP‐G, deletion of the cTP (chloroplastic transit peptide), ORF fused to GFP; ΔΔ‐G combined deletion of NLS and cTP in the ORF fused to GFP. The variants are expressed under the control of the CaMV35S promoter or under the native PAP8‐1.1-kb promoter. Plasmid identification and relevant restriction sites for cloning are given in light grey (see Appendix Table S1); mcs, multiple cloning site.
-
BTransgenic lines obtained with the constructions described above. Three phenotypic classes have been recorded corresponding to albino (white squares), pale green (light green squares) and green plants (green squares). Col‐0, wild type; pap8‐1/+, heterozygous mixture; n, total number of recorded plants; ø, no transgene.
-
CFunctional complementation output. Hygromycin‐resistant plants were transferred on soil and grown under long‐day conditions (16‐h light/8-h dark; ˜ 70 μmol m−2 s−1) at 21°C and 60% humidity. Genomic DNA was isolated from true leaves and used for genotyping. The presence of the pap8‐1 allele was confirmed using the primer ortpF/oLBb1.3. PAP8 wild‐type allele tested with ortpF/op8i2_R. The insertion of the transgene of interest was tested with oPAP8_rtp_F/oE3_R. The number of the tested plants (HygR#T1), the number of double heterozygous plants (p8‐1/+; TG/−) and the number of sesqui‐mutant plants (p8‐1/p8-1; TG/−) are depicted for each construction; p8P8 presented as positive control. None of the 39 T1 plants with pP8::PAP8‐GFP were photosynthetic and homozygous pap8‐1. Therefore, two doubly heterozygous (pap8‐1/+; TG/−) expressing GFP were tested for their segregation pattern (Line 1: 34% albino, n = 169, and Line 2: 24% albino, n = 199) and compared to that of pap8‐1/+ (28% albinos, n = 99). In the absence of statistical difference between the samples (ε = 0.102≪1.96 for α = 0.05; ε‐test, Fisher Yates), PAP8‐GFP was declared not functional as opposed to PAP8.
Source data are available online for this figure.
