Figure 3.
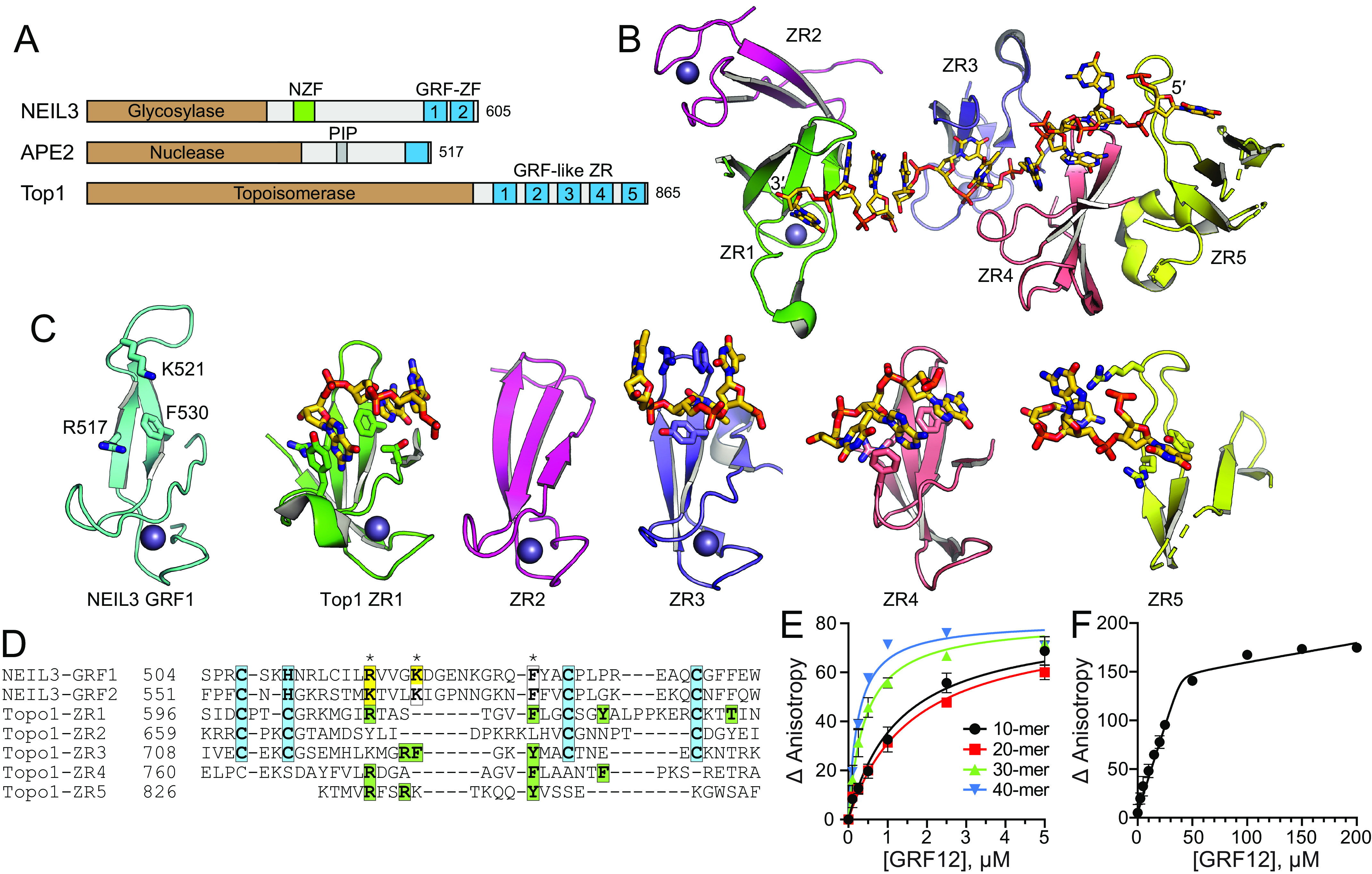
DNA-binding modes by GRF motifs. A, NEIL3, APE2, and E. coli TopI contain GRF–ZF and GRF-like ZR motifs at their C terminus. B, the structure of the TopI ZR domain, with DNA shown as sticks. C, comparison of hNEIL3 GRF1 (cyan) with individual TopI ZR motifs (colored as in B). DNA-binding residues and DNA are shown as sticks. D, structure-based sequence alignment. Zn2+-coordinating residues are highlighted cyan. NEIL3 residues that affect DNA binding upon mutation are highlighted yellow, and DNA interacting residues in the TopI structure are green. Asterisks mark the highest conservation among DNA-binding positions. E, DNA binding of mNEIL3 GRF12 to ssDNA of varying lengths. Total [DNA] used was 25 nm. The data are means ± S.D. (n = 3). F, stoichiometry of binding of GRF12 to 40-mer ssDNA. Total [DNA] = 5.05 μm (≫ Kd). The inflection point in the titration curve fits to [GRF12] = 40 μm, which equals an 8:1 GRF12:DNA molar ratio.
