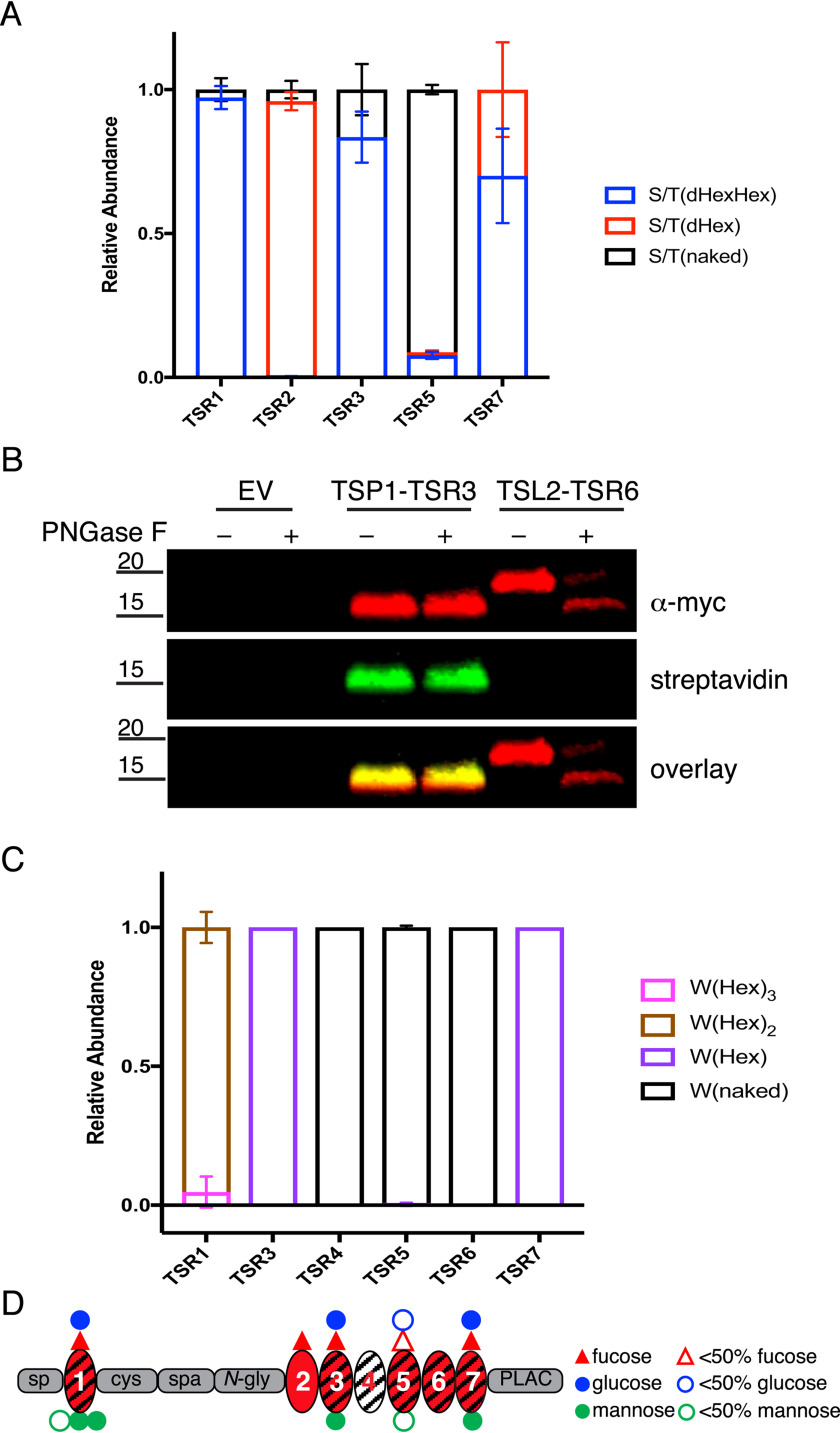
Figure 2.
ADAMTSL2 TSRs were modified with O-fucose-glucose disaccharide and C-mannose in varying stoichiometries. A, relative abundance of GlcFuc disaccharide, O-fuc monosaccharide, and unmodified glycoforms on peptides from TSRs containing the C-X-X-(S/T)-C O-fucose consensus site analyzed by mass spectral analyses. See Fig. S2 for spectra and EICs of each peptide. Data can be found in Tables S3, S4, S7, and S8. B, plasmids encoding TSP1-TSR3-MycHis6, ADAMTSL2-TSR6-MycHis6, or an empty vector (EV) were transfected into HEK293T cells grown in the presence of 6AF as described under “Experimental procedures.” The proteins were purified from the medium, digested with or without PNGase F, click reaction was performed with azide-biotin, separated by SDS-PAGE, and probed with anti-myc antibody (red) or streptavidin (green). C, relative abundance of C-mannosylated forms of peptides from TSRs containing W-X-X-(W/C) consensus sequence. See Fig. S2 for spectra and EICs of each peptide. Data can be found in Tables S3, S4, S7, and S8. D, domain map of mADAMTSL2 summarizing the distribution of O-fucosylation and C-mannosylation on TSRs. Red triangle represents fucose. Blue circle represents glucose. Green circle represent mannose. Hollow icons represent less than 50% modification of fucose (red hollow triangle), glucose (blue hollow circle), and mannose (green hollow circle).