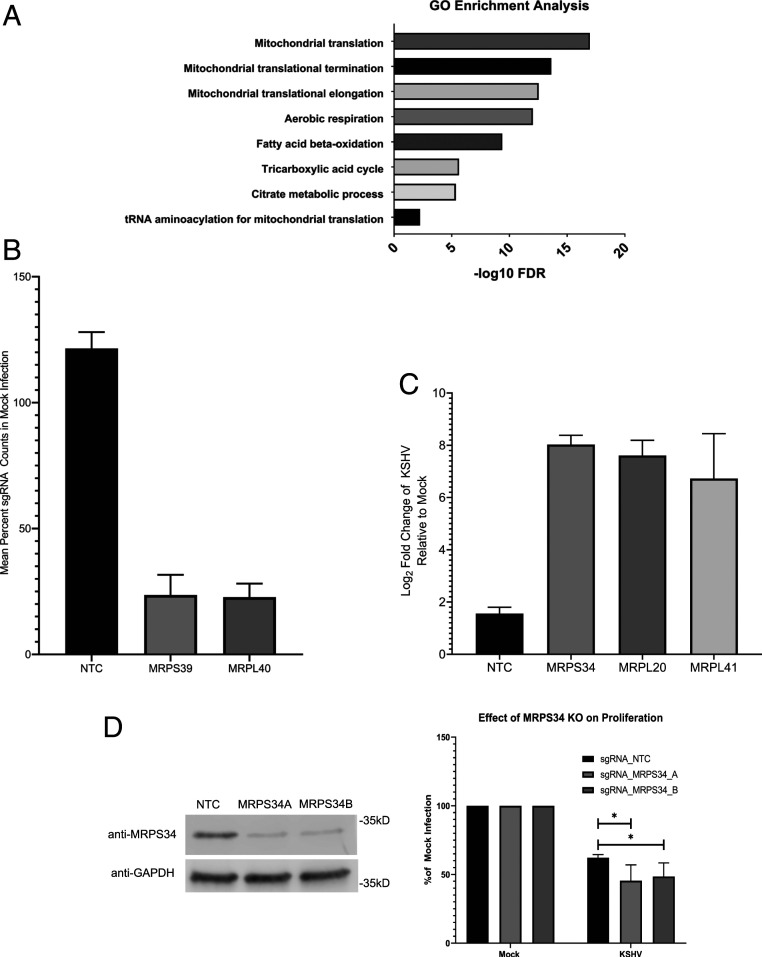
Fig. 2.
Genes involved in mitochondrial translation are essential during KSHV infection. (A) Gene ontology (GO) enrichment analysis of the hits that localize to the mitochondria indicate that mitochondrial translation is a common function for many of these genes. FDR, false discovery rate. (B) The sgRNA counts from the live cell screen for two components of the mitochondrial ribosome, MRPS39 and MRPL40, are depleted relative to counts in uninfected cells when compared with nontargeting controls. (C) sgRNA counts from the dead cell screen for three components of the mitochondrial ribosome are enriched in the KSHV-infected cells relative to the uninfected controls when compared with the nontargeting controls. (D) Western blot showing suppression of protein expression with sgRNAs targeting MRPS34 as well as a nontargeting control (NTC) sgRNA. The separate sgRNAs targeting MRPS34 are able to knock out (KO) the gene in a portion of transduced TIME cells and significantly reduce cell growth relative to mock infection when compared with nontargeting control transduced cells (paired t test; n = 4). *P < 0.05.