Fig. 3.
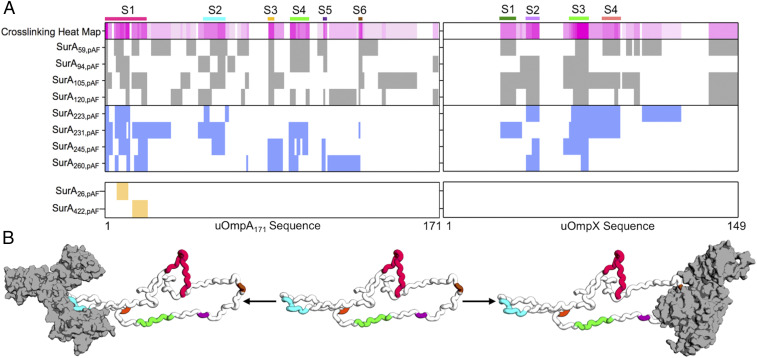
Photo-crosslinking mass spectrometry (pXL-MS) identifies segments on client uOMPs that bind SurA. (A) The crosslinking patterns for eight high-efficiency SurApAF variants are shown to two uOMP clients (uOmpA171 and uOmpX). Constructs that place pAF on the core domain are colored gray (top register), and constructs that place pAF on the P1 domain are colored blue (second register). The crosslinking heat map depicts the frequency a given residue on a client uOMP crosslinks to pAF in eight separate crosslinking experiments (darker magenta indicates residue is crosslinked more often). Binding segments are demarcated with a colored bar above the heat map and a label (S1 to 6 or S1 to 4). The bottom register shows results from two negative control studies using SurA26,pAF and SurA422,pAF (see main text for explanation). Only one uOmpA171 crosslinked peptide was found for each of these constructs while uOmpX did not crosslink at all. (B) An expanded uOmpA171 model with hydrodynamic properties consistent with the contrast-matched SANS experiment is shown as a schematic with the SurA-binding segments colored as in A. Two different segments (S2, left structure, o1s022; and S6, right structure, o1s021) are shown bound to SurA (shown in gray with a surface representation), suggesting that more than one copy of SurA could bind a single copy of uOmpA171 with minimal steric clash.