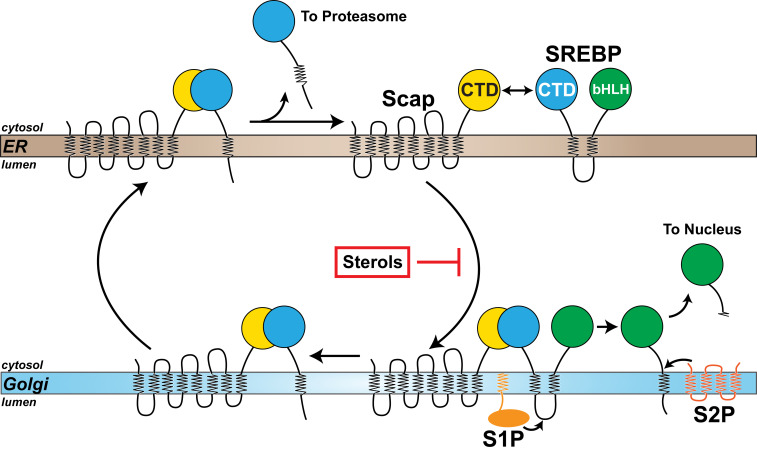
Fig. 9.
A model of the SREBP pathway. SREBP is comprised of a large NTD (green) and a large CTD (cyan), both of which project into the cytosol and are anchored to the ER membrane by two transmembrane helices connected by a short luminal loop. The NTD contains a basic helix–loop–helix leucine zipper transcription factor domain (bHLH) and the CTD binds to the cytosol-facing CTD of Scap (yellow), a cholesterol-sensing membrane protein containing eight transmembrane helices. When cellular cholesterol levels fall below a threshold, Scap escorts SREBP from the ER to the Golgi. Transport is blocked when sterols accumulate. Upon arrival at the Golgi, SREBP is sequentially cleaved by two Golgi-resident proteases, site-1 protease (S1P) and site-2 protease (S2P), releasing its NTD from the membrane for translocation to the nucleus to up-regulate expression of lipogenic genes. Following cleavage by S1P, the CTD of SREBP remains bound to Scap and returns to the ER with Scap. Once in the ER, the CTD of SREBP dissociates from Scap and is degraded by the proteasome, liberating Scap to bind to additional SREBPs and undergo transport to the Golgi if cellular cholesterol levels are still below a threshold set point.