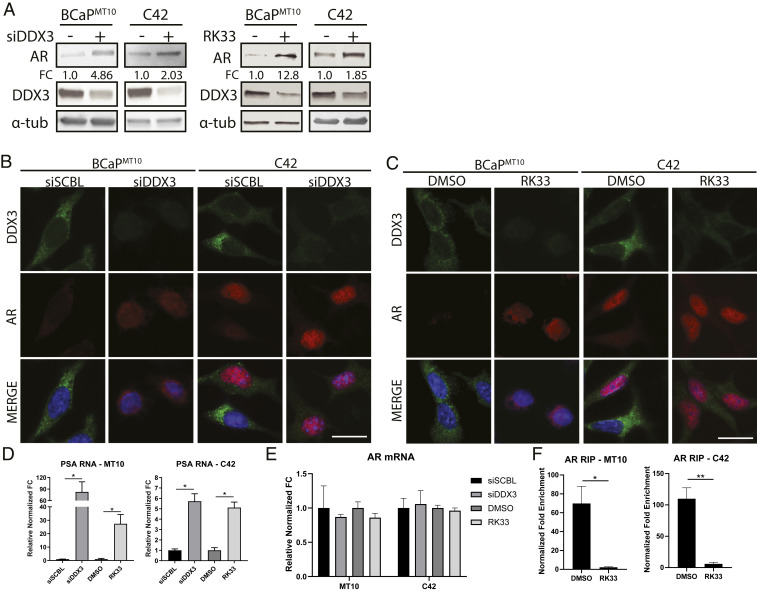
Fig. 4.
Genetic and pharmacological inhibition of DDX3 increases AR protein expression and signaling. Inhibition of DDX3 resulted in increased AR protein expression and AR signaling in CRPC. (A) Western blot analysis of AR protein expression in BCaPMT10 and C42 bulk populations after inhibition of DDX3 by siRNA and small-molecule inhibitor RK33 showed an increase of overall AR protein and decrease of DDX3 protein. With siRNA, AR protein increased 4.86-fold in BCaPMT10 and 2.03-fold in C42 compared to scramble control. With RK33 treatment, AR protein increased 12.8-fold in BCaPMT10 and 1.85-fold in C42 compared to DMSO control. α-tub was used as a loading control. (B) Immunofluorescence analysis of DDX3 expression and localization after inhibition with siRNAs showed decreased cytoplasmic DDX3 expression (green) and increased AR expression (red), compared to scramble control (siSCBL) in two CRPC models (BCaPMT10 and C42). Nuclei were counterstained with DAPI (blue). (C) Immunofluorescence analysis of DDX3 expression and localization after pharmacologic inhibition using 2 µM RK33 showed decreased cytoplasmic DDX3 expression (green) and increased AR expression (red) compared to DMSO control in BCaPMT10 and C42. Nuclei were counterstained with DAPI (blue). (D) Assessment of AR signaling, using PSA, a transcriptional target of AR, was significantly increased with siDDX3 compared to siSCBL (BCaPMT10, P = 0.036; C42, P = 0.021) and with RK33 compared to DMSO (BCaPMT10, P = 0.019; C42, P = 0.029) in both CRPC models. (E) qPCR for AR mRNA showed no significant difference in expression between siSCBL vs. siDDX3 and RK33 vs. DMSO. (F) RIP analysis of AR mRNA bound to DDX3 after RK33 treatment (RK33) showed a significant decrease in DDX3 binding to AR mRNA compared to control (DMSO) in CRPC cell lines BCaPMT10 (P = 0.02) and C42 (P = 0.004). (Scale bars, 10 µm.) Bar graphs represent mean ± SEM. Significance is represented by *P ≤ 0.05, **P ≤ 0.01.