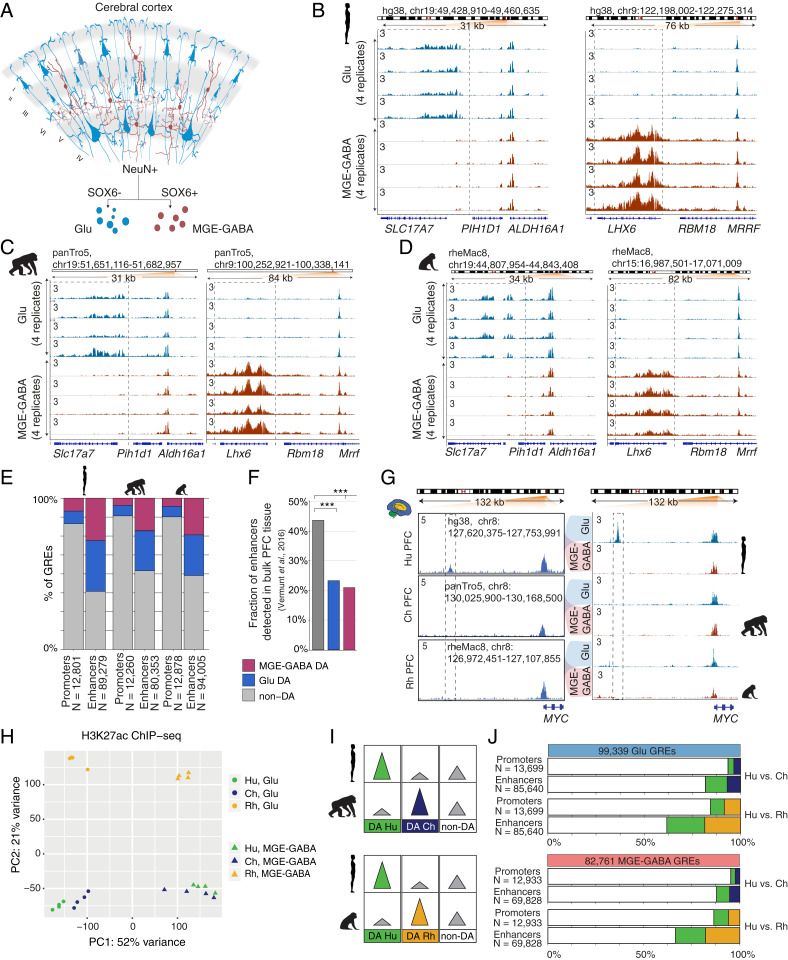
Fig. 1.
Regulatory changes in Glu and MGE-GABA neurons during primate evolution. (A) Schematic of isolation of Glu and MGE-GABA nuclei. (B–D) H3K27ac ChIP-seq profiles for Hu (B), Ch (C), and Rh (D) at the loci of Glu (SLC17A7, Left) and MGE-GABA (LHX6, Right) markers. Read per million (RPM)-normalized reads (axis limit 3 RPM). (E) Fractions of promoters or enhancers that are Glu DA, MGE-GABA DA or non-DA in Glu vs. MGE-GABA. (F) Fractions of Glu DA, MGE-GABA DA, and non-DA Hu enhancers that overlap GREs in bulk prefrontal cortex (PFC) tissue (12). ***P < 0.0005 (Fisher’s exact test). (G) The regulatory landscapes near the MYC locus in bulk PFC (12) (Left) and in Glu and MGE-GABA neurons (Right). (H) PCA of ChIP-seq data for Glu and MGE-GABA neurons from Hu, Ch, and Rh. (I) Schematic of pairwise interspecies comparisons between the ChIP-seq datasets. (J) Fractions of Glu DA, MGE-GABA DA, and non-DA GREs in pairwise species comparisons; Glu (Upper) and MGE-GABA (Lower) neurons.