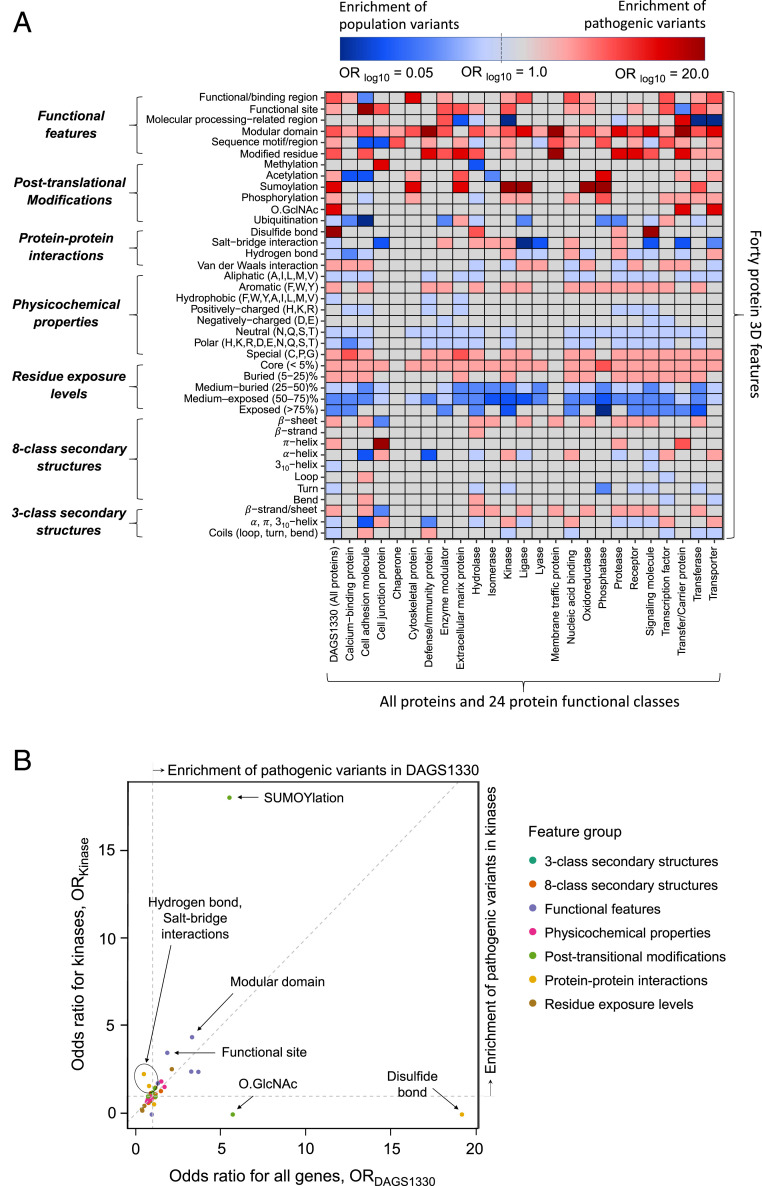
Fig. 3.
Some features of 3D mutational hotspots are conserved across different protein functional classes, whereas others are unique to specific classes. (A) Heatmap of ORs found from the burden analyses (two-sided Fisher’s exact test) on 40 3D features with pathogenic and population variants from all 1,330 disease-associated genes (full DAGS1330 dataset) and for subsets of genes grouped into 24 protein classes based on their molecular functions. To facilitate the visualization, minimum and maximum values of OR are set to 0.05 and 20.0, respectively. The red and the blue color gradients represent different degrees of association to pathogenic (1.0 OR 20.0 and 0.05) and population (0.05 OR 1.0 and 0.05) variants; darker color indicates stronger association. The gray cells in the heatmap represent features that are not significantly associated ( 0.05) with any variation type. Thus, the rows with only red or blue cells show the characteristic features of pathogenic or population variations that are consistent or conserved across all of the protein classes. In contrast, the rows with both red and blue cells indicate protein class-specific diverging features. (B) Scatter plot showing the correlation between the burden of pathogenic variations on different features for all genes along the x axis () and for kinase protein class along the y axis (). Each circle represents a protein feature (indicated by an arrow), and has a different color according to the seven main feature categories. The diagonal line represents the agreement between the burden values found for all genes and those for kinases. The features above the diagonal line and to the left of the vertical line are enriched with pathogenic variations in kinases (hydrogen bond and salt bridge interaction sites), but are depleted of pathogenic variations in the full DAGS1330 set. The features above the diagonal line and to the right of the vertical line have an elevated burden of pathogenic variations in kinases (y axis), indicating that these features are more intolerant to substitutions for this protein class compared to the general trend for all proteins (x axis). In contrast, the features below the diagonal line and the horizontal line are enriched with pathogenic variations in the DAGS1330 set (disulfide bond and O.GlcNAc), but are depleted of pathogenic variations in kinases.