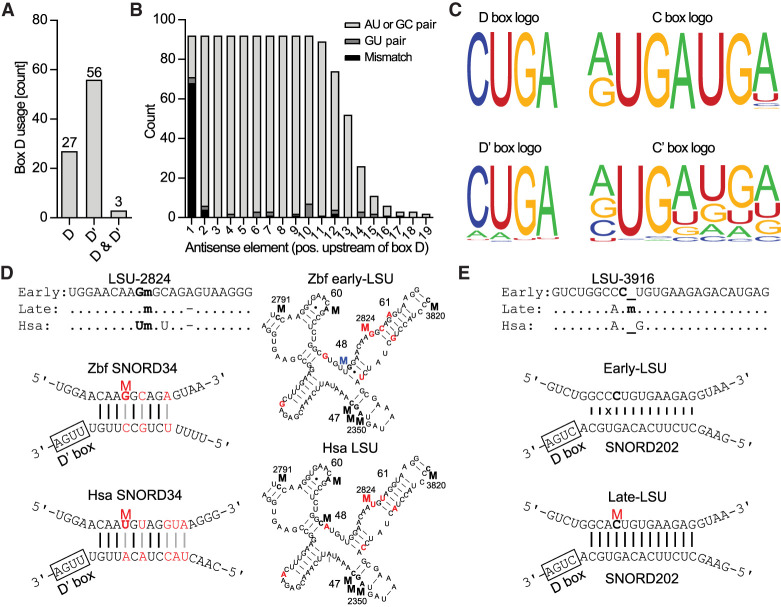
FIGURE 4.
Analysis of SNORDs identified in this study. (A) Distribution of SNORDs guiding methylations in rRNA using box D, box D′, or both. (B) Base pair interaction between the antisense element in the SNORD and the rRNA target in relation to the distance from box D (or D′). (C) Sequence logo of the box D, D′, C, and C′ of SNORDs guiding methylations in rRNA generated by WebLogo (Crooks et al. 2004). (D, upper left) sequence comparison of human and zebrafish early- and late-rRNA containing a conserved methylation at LSU-2824 with indications of identical (.), deleted (-), and methylated (m) nucleotides. (Lower left) Human and zebrafish SNORD34 and the interaction with the target sequence surrounding LSU-2824. Nucleotides that differ between the two are labeled in red. (Right) 2D structures of a conserved LSU rRNA region between human and zebrafish highlighting methylations in H47, H48, H60, and H61. Methylation at LSU-2824 is in red and the zebrafish-specific methylation in blue. (E, upper) sequence comparison of human and zebrafish early- and late-rRNA highlighting LSU-Cm3916 only found in late-rRNA. (Lower) Drawings of the SNORD base-pairings with early- and late-rRNA, respectively. The presence of a mismatch (x) with early-rRNA may prevent pairing and methylation. (Hsa) Homo sapiens, (Zbf) zebrafish.