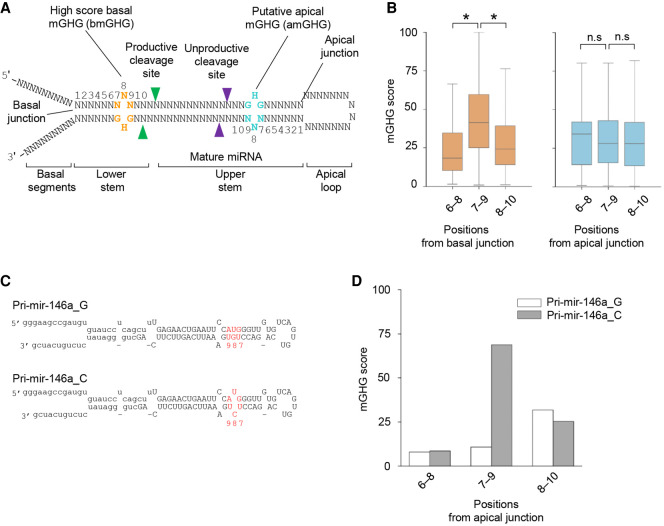
FIGURE 3.
The SNP G/C generates a noncanonical apical mGHG motif in the apical region. (A) The positions of the canonical mGHG motifs at the basal junction in pri-miRNAs. The mGHG motif contains a mismatch at the eighth nucleotide from the basal junction. A putative apical mGHG motif might occur at the apical junction. (B) The mGHG scores were estimated for the mGHG motifs at the apical and basal junctions. Statistically significant and nonsignificant differences between the various data sets are indicated by an asterisk (*) and n.s., respectively (two-tailed t-test; mGHG scores of positions 6–8 vs. positions 7–9 from basal junction: P = 1.96 × 10−50, positions 7–9 vs. position 8–10 from basal junction: P = 3.83 × 10−34). (C) The putative apical mGHG motif in pri-mir-146a_C is shown in red. (D) mGHG scores of the mGHG motifs in positions 6 to 10 at the apical junction of pri-mir-146a_G or C.