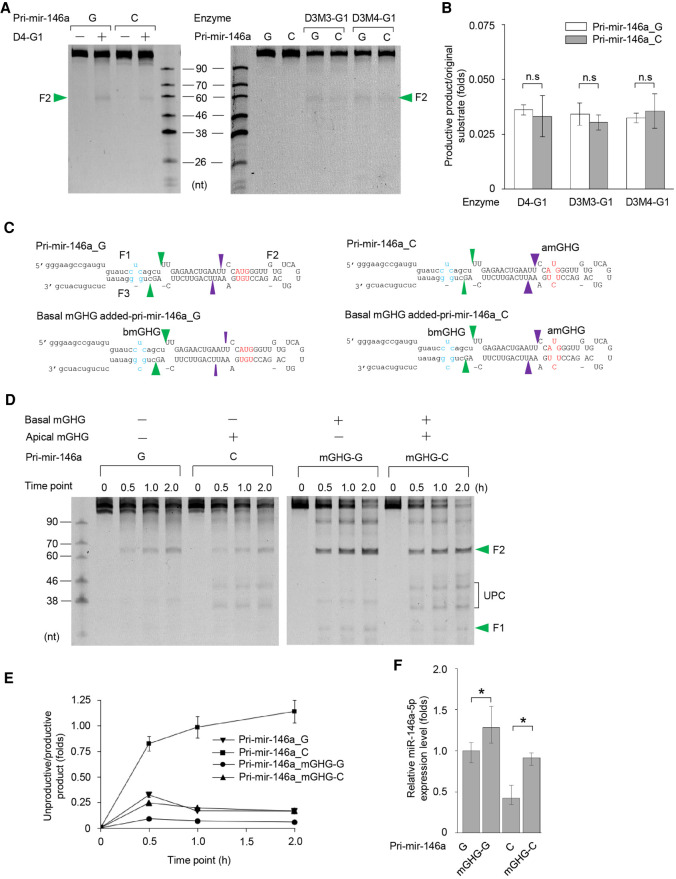
FIGURE 4.
The apical mGHG motif recruits DROSHA to the apical junction via interaction with its dsRBD. (A) The cleavage of pri-mir-146a_G and C by mutant dsRBD-containing DROSHA. One pmol of each RNA was incubated with 6 pmol D3M3–G1, 6 pmol D3M4–G1, or 30 pmol D4–G1 in 10 µL pri-miRNA processing buffer. The green arrowheads indicate the productive cleavages. (B) The cleavage activity of the D3–G1 complexes (as shown in A) was calculated by estimating the ratio of the productive product to the original substrate from three independent experiments. No significant (n.s.) differences were found between the two data sets (two-tailed t-test). (C) The added basal mGHG motifs in pri-mir-146a_G and C are shown in blue. (D) Cleavage of the basal mGHG-added pri-mir-146a_G and C by Microprocessor (NLSD3–DGCR8). Five pmol of each RNA was incubated with 5 pmol Microprocessor in 10 µL pri-miRNA processing buffer for 0.5, 1, and 2 h. UPC indicates the unproductive products, whereas the green arrowheads indicate productive products. (E) The unproductive cleavage of Microprocessor (shown in D) was assessed from three independent experiments. (F) The qPCR-estimated miR-146a expression levels in human cells transfected with pcDNA3 plasmid expressing each of four pri-mir-146a in D. Statistically significant and nonsignificant differences between the various data sets are indicated by an asterisk (*) and n.s., respectively (two-tailed t-test; relative miR-146a expression level of pri-mir-146a_G vs. pri-mir-146a_mGHG-G: P = 0.043, pri-mir-146a_C vs. pri-mir-146a_mGHG-C: P = 0.029).