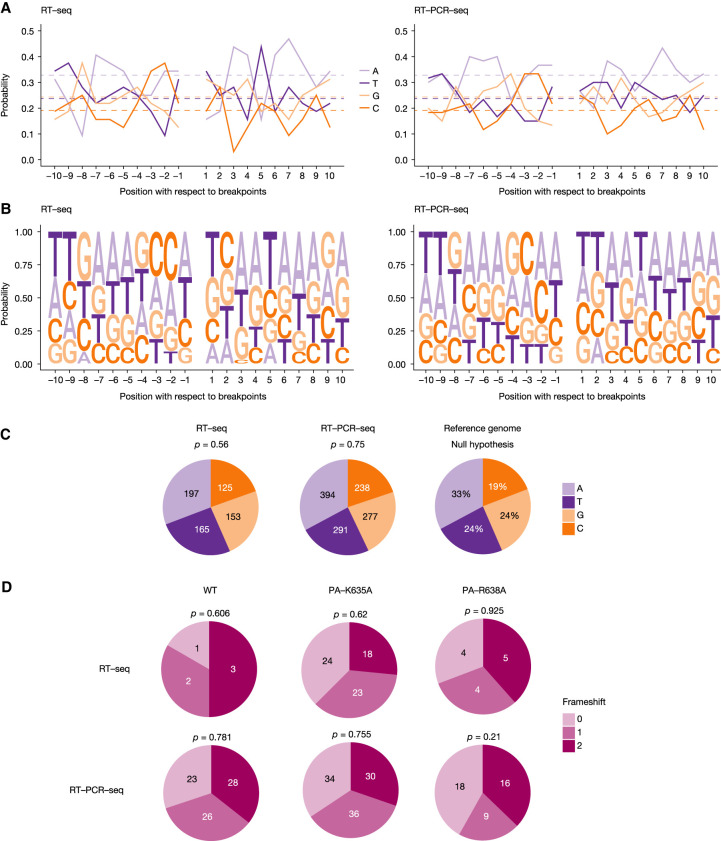
FIGURE 5.
DVG breakpoints. (A–C) Nucleotidic composition of the sequences adjacent to the DVG breakpoints. Analysis was performed by combining DVGs obtained from all segments for the WT and mutant viruses. (A,B) The frequency of each nucleotide found both upstream and downstream from the breakpoints is shown. Negative and positive numbers denote positions upstream of the breakpoint start and downstream from the breakpoint end, respectively. As a reference, the overall frequency of each nucleotide in the IAV genome is indicated by the dashed horizontal lines. (C) Distribution of each nucleotide when grouping positions −10 through +10, compared with the null distribution consisting of IAV reference sequences through all segments. Significance was determined using χ2 goodness-of-fit test. (D) Pie charts showing the relative proportion of DVGs found with each frameshift, depending on the method used. Significance was determined using χ2 goodness-of-fit test.