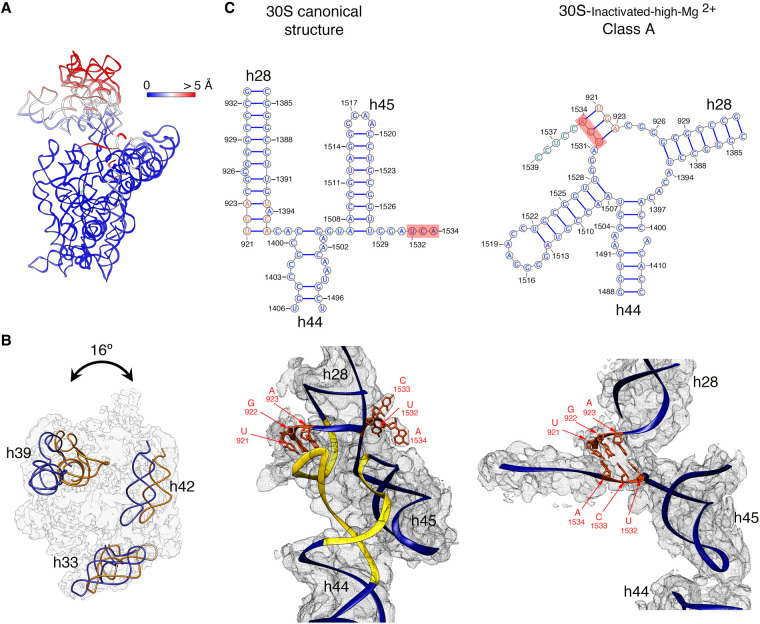
FIGURE 2.
Molecular model of the 30S-inactivated-high-Mg2+ particle. (A) Temperature map of the 30S-inactivated-high-Mg2+ class A. The rRNA is colored according to the r.m.s.d. deviation (Å) with respect to the structure of the 30S subunit obtained by X-ray crystallography (PDB ID 4V4Q). (B) Top view of the head of the cryo-EM map of the 30S-inactivated-high-Mg2+ class A. The positions of helices 33, 39, and 42 in this structure (navy blue) and in the crystal structure of the 30S subunit (orange) are shown to illustrate the backward tilting of the head by 16° in the structure of the 30S-inactivated-high-Mg2+ class A. (C) Secondary (top panels) and tertiary (bottom panels) structures of helices 28, 44, and 45 of the 16S rRNA in the 30S subunit structure obtained by X-ray crystallography and in the molecular model derived from the cryo-EM map of the 30S-inactivated-high-Mg2+ class A. The nucleotides 1532–1534 at the 3′ end of the 16S rRNA involved in the conformational transition between both structures are highlighted in red. The regions of helices 44 and 28 that become unfolded during the conformational transition are colored in yellow in the canonical 3D structure of the 30S subunit. The molecular model of the cryo-EM map of the 30S-inactivated-high-Mg2+ class A is shown overlapped with the density of the obtained cryo-EM map. In the case of the structure of the 30S subunit obtained by X-ray crystallography, a density map was generated from the atomic coordinates and overlapped with the molecular model.