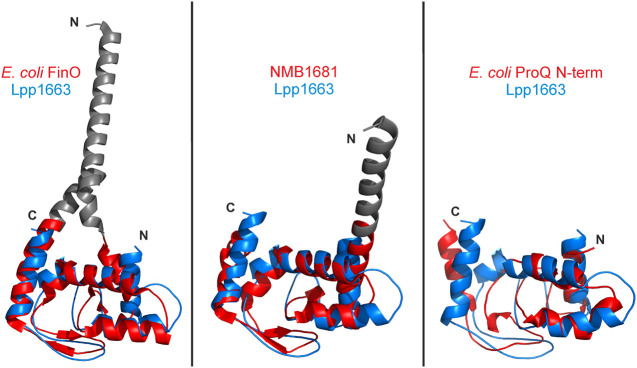
FIGURE 3.
The ProQ/FinO fold is well conserved. The lowest energy Lpp1663 NMR solution structure (blue) was superposed with the FinO/ProQ domains of E. coli FinO (PDB:1DVO), N. meningitidis NMB1681 (PDB:3MW6), and E. coli ProQ (PDB:5NB9) colored in red. The Cα-rmsd between the structures are 1.8 Å (80 atom pairs), 1.9 Å (91 atom pairs), and 4.2 Å (75 atom pairs), respectively, which shows the high degree of conservation of the core fold for the ProQ/FinO domain. The rmsd was calculated with the alignment tool in PyMol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). Flexible amino- and carboxyl termini are not shown and loops were smoothed for clarity. Amino- and carboxyl termini are indicated as N and C, respectively.