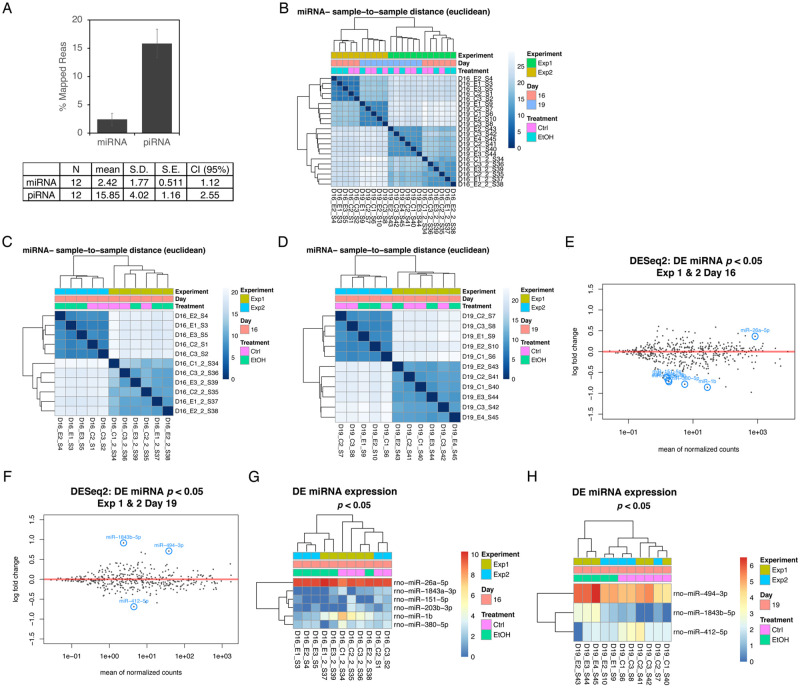
Fig 2. Bioinformatics analysis of miRNA-Seq data on amniotic exosomal miRNAs.
(A) Processed RNA-seq data was mapped and found piRNA enriched in rat amniotic fluids compared to miRNA. Error bars represent 95% confidence interval. (B) Dendrogram was built to examine correlation among miRNAs in samples depending on the experiment (Exp1 and 2), collection time points (E16 and E19) and treatment (control vs. EtOH). Correlation among AF exosomal miRNAs for (C) E16 samples from Exp1 and 2 and (D) E19 samples from Exp1 and 2. (E) DESeq2 differential expression analysis for E16 samples from Exp1 and 2. (F) DESeq2 differential expression analysis for E19 samples from Exp1 and 2. (G) Selected six miRNAs with p < 0.05 differentially expressed in E16 samples from Exp1 and 2 are shown with circle. (H) Selected three miRNAs with p < 0.05 differentially expressed in E19 samples from Exp1 and 2 are shown with circle.