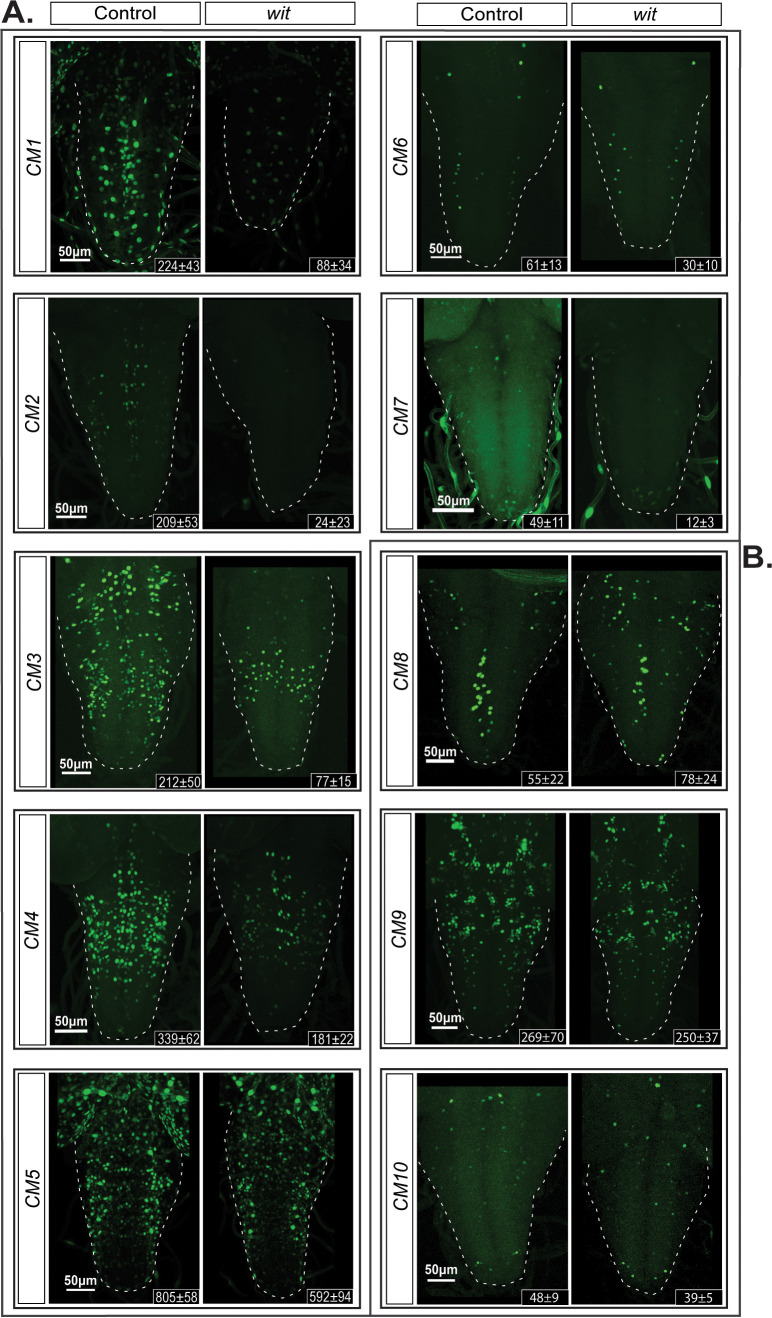
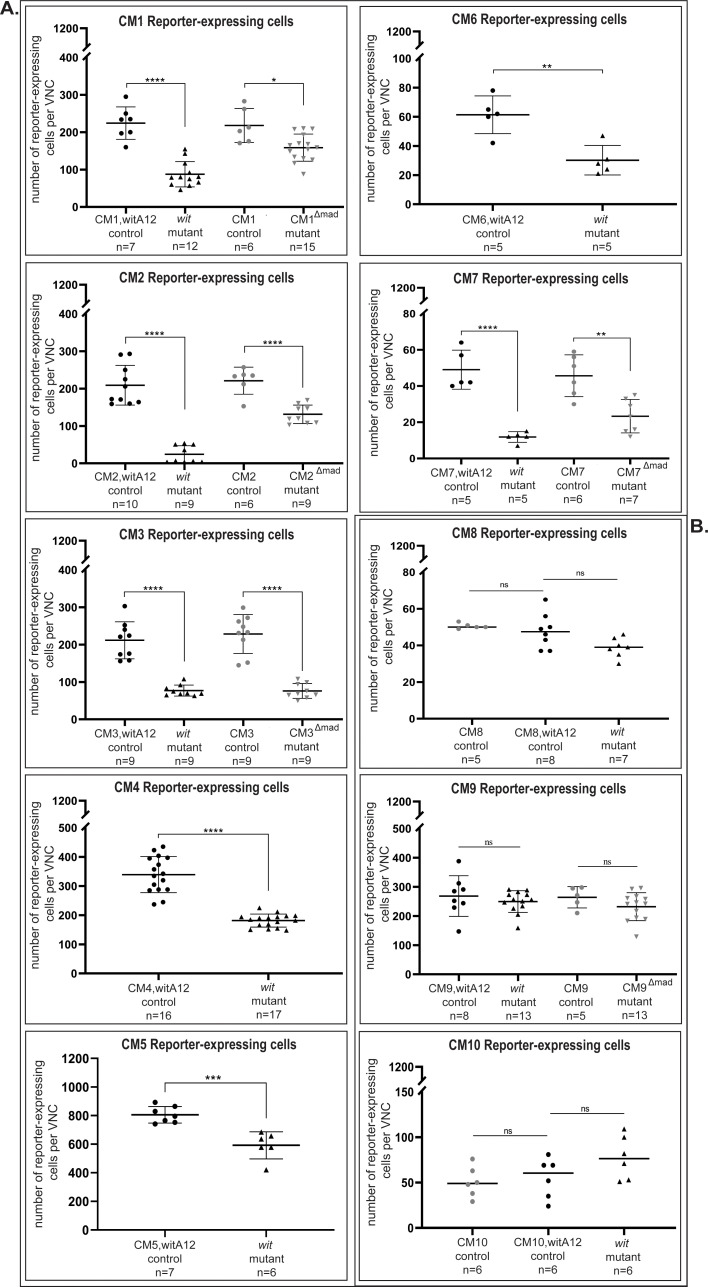
(A) None of the three reporter fragments (CM8, CM9, CM10) showed a significant change in number of reporters expressing cells between controls and their respective mutants. (B) In the VNC, the CM1 (CM1,witA12/+ controls) reporter was expressed in 224 ± 43 nuclei, of which 56 ± 16 (25%) nuclei are pMad-positive (n = 7). In wit mutants (witA12/witB11), reporter activity was reduced by 61% to 88 ± 34 nuclei (n = 12). Once the pMad-binding site was mutated (CM1Δmad), reporter activity was reduced by 27% to 159 ± 36 nuclei (n = 15) compared to controls that had reporter expression in 218 ± 46 nuclei (n = 6). CM2 (CM2,witA12/+ controls) reporter was expressed in 209 ± 53 nuclei, of which 111 ± 33 (53%) nuclei are pMad-positive (n = 10). In wit mutants (witA12/witB11), reporter activity was reduced by 88.5% to 24 ± 23 nuclei (n = 9). In the pMad-binding site mutant (CM2Δmad), reporter activity was reduced by 41% to 131 ± 25 nuclei (n = 9) compared to controls that had reporter expression in 221 ± 36 nuclei (n = 6). CM3 (CM3,witA12/+ controls) reporter was expressed in 212 ± 50 nuclei, of which 68 ± 19 (32%) nuclei are pMad-positive (n = 9). In wit mutants, reporter activity was reduced by 64% to 77 ± 15 nuclei (n = 9). In the pMad-binding site mutant (CM3Δmad), reporter activity was reduced by 67% to 76 ± 20 nuclei (n = 9) compared to controls that had reporter expression in 228 ± 52 nuclei (n = 9). CM4 reporter was expressed in 339 ± 62 nuclei, of which 134 ± 25 (34%) nuclei were pMad-positive (n = 16). In wit mutants, reporter activity was reduced by 47% to 181 ± 22 nuclei (n = 17). CM5 reporter was expressed in 805 ± 58 nuclei in the VNC, of which 217 ± 10 (27%) nuclei were pMad-positive (n = 7). In wit mutants, reporter activity was reduced by 30% to 592 ± 94 nuclei (n = 6). CM6 was expressed in 61 ± 13 nuclei (n = 5). In wit mutants, reporter activity was reduced by 50% to 30 ± 10 nuclei (n = 5). Finally, CM7 (CM7,witA12/+ controls) reporter was expressed in 49 ± 11 nuclei, of which 9 ± 2 (18%) nuclei are pMad-positive (n = 5). In wit mutants, reporter activity was reduced by 75% to 12 ± 3 nuclei (n = 5). In the pMad-binding site mutant (CM7Δmad), reporter activity was reduced by 50% to 23 ± 9 nuclei (n = 7) compared to controls that had reporter expression in 46 ± 12 nuclei (n = 6). Significance was calculated with One-way ANOVA with a Tukey post hoc multiple comparisons test for the following genotypes: CM1 control versus wit mutants p<0.0001 and control versus pMad-binding site mutants p=0.0143; CM2 control versus wit mutant and control versus pMad-binding site mutants p<0.0001; CM7 control versus wit mutants p<0.0001 and control versus pMad-binding site mutants p=0.0021. Student`s t-test was used for: CM4 p<0.0001; CM5 p=0.0004; CM6 p=0.0028. For CM3, as the wit control samples were non-normally distributed (Shapiro Wilk test), significance was calculated with the Mann-Whitney U-test for control versus wit mutants p<0.0001; Student's t-test was used for control versus pMad-binding site mutants p<0.0001. Each point represents the total number of reporter-expressing cells in the VNC of a single animal, n indicates the number of VNCs analyzed and data is reported as mean ± SD. Genotypes: All control and pMad-binding site mutant lines examined here were heterozygous (w;;CM#/+); wit mutants (w;;CM#,witA12/witB11).