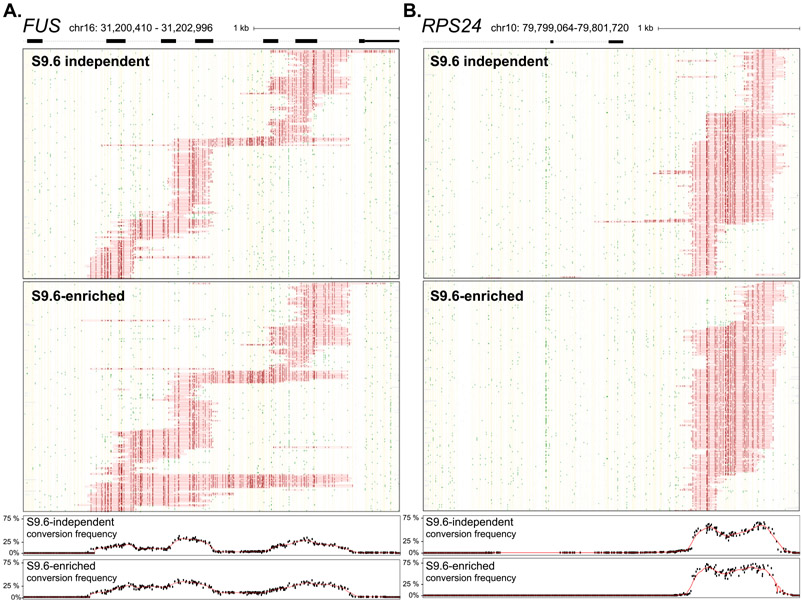
Fig. 2. Comparison of native and DRIP-enriched SMRF-seq profiles.
(A) SMRF-seq profiles are shown for the nontemplate strand of the FUS amplicon without and with S.6 enrichment (top and bottom, respectively). Two hundred twenty-six molecules are shown in both panels corresponding to the totality of footprints in the absence of S9.6 and to a random subsample after S9.6 enrichment. The plots below depict the aggregate C to T conversion frequencies observed for each sample over R-loop-containing molecules. (B) Similar data as in previous panel for the RPS24 locus. Two hundred fifty-eight independent molecules are shown in both panels corresponding to the totality of footprints in the absence of S9.6 and to a random subsample after S9.6 enrichment.