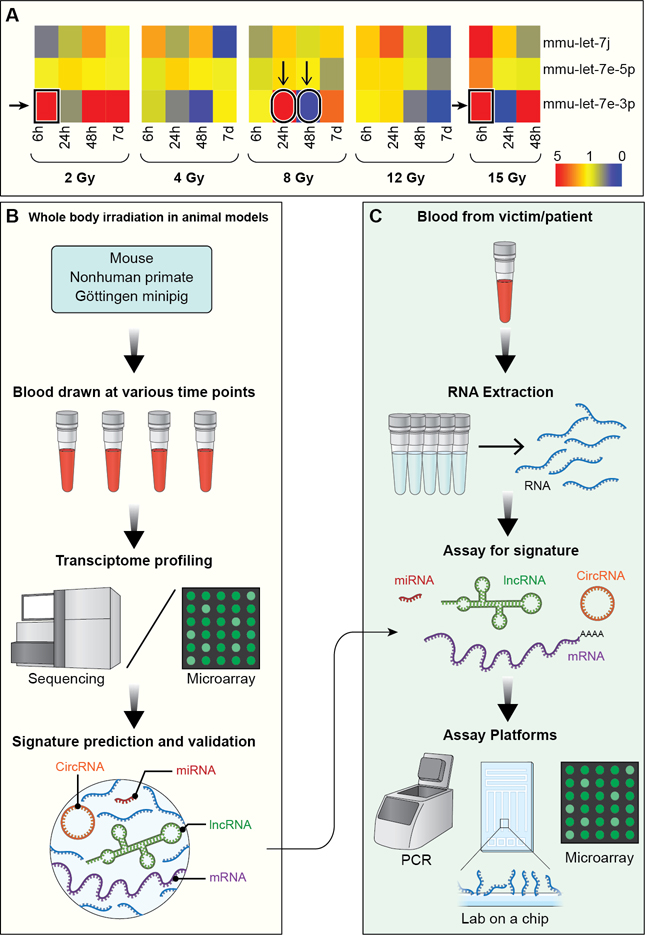
Figure 4: Predicting radiation exposure and normal tissue injury using an integrated RNA biomarker approach.
A) Heatmap displays fold change values of 3 different miRNAs of let-7 family, which share a seed sequence indicating shared targets, expressed in the blood of mice collected at various timepoints after whole body irradiation with various doses. Red signifies upregulated expression and blue signifies downregulated expression in comparison to the sham-irradiated controls. B) Schematic shows the methodology of utilizing animal models to predict dose-differentiating, RNA-based signatures from approximately 300–500μL of blood. C) Intended workflow, starting with a 500μL blood draw and using different high throughput diagnostic platforms, shows application of predicted signatures for triaging victims of accidental radiation exposure and assessment of normal tissue injury after radiotherapy.