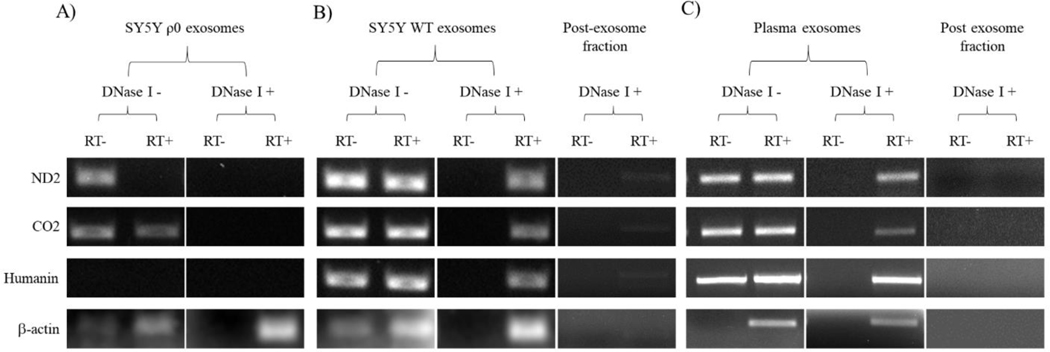
Figure 8. Interrogation of exosome fraction RNA samples for mtDNA-derived mRNA transcripts.
(A) RNA from SH-SY5Y ρ0 cell-derived exosome fractions, with or without post-isolation DNase I treatment, was subjected to amplification with primers to mtDNA-derived mRNA transcripts (ND2, CO2, and humanin). Primers to β-actin, a nuclear gene-derived transcript, were also utilized. The PCR reactions were performed without prior reverse transcription of the RNA to cDNA (RT-) and following reverse transcription of the RNA to cDNA (RT+), and the resulting PCR products were analyzed using gel electrophoresis. (B) An equivalent analysis is shown for RNA from SH-SY5Y cell-derived exosome fractions. In addition, we analyzed RNA isolated from the post-exosome fractions. (C) An equivalent analysis is shown for RNA from plasma-derived exosome and post-exosome fractions. For all cases the data shown are representative of three experiments.