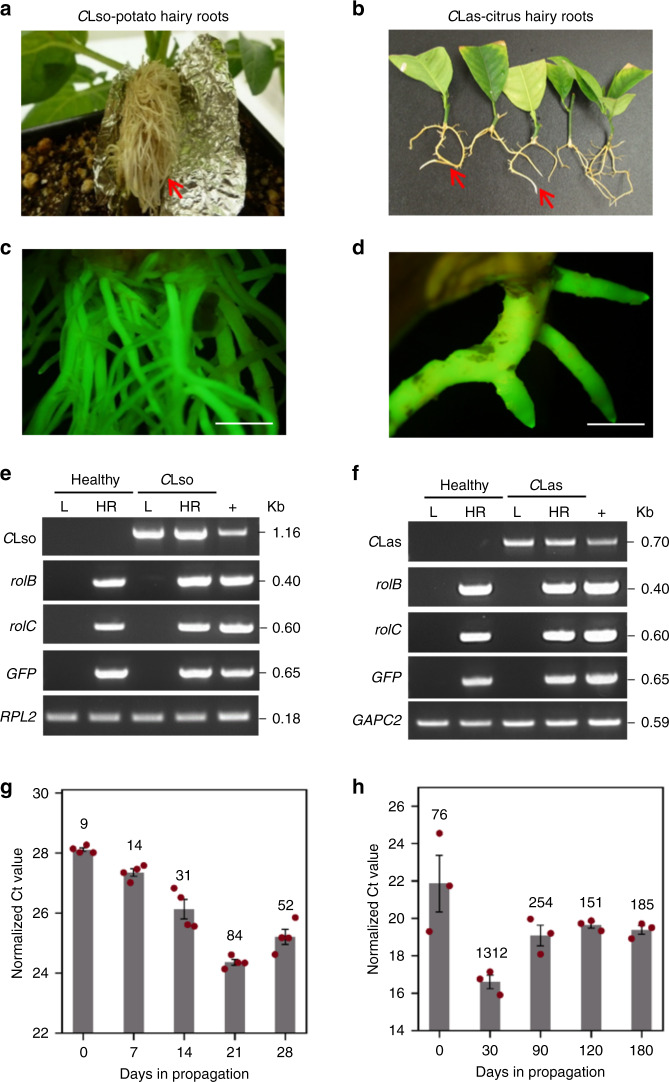
Fig. 1. Candidatus Liberibacter solanacearum and Candidatus Liberibacter asiaticus in potato and citrus hairy roots.
a, b Hairy roots (indicated by arrows) from Candidatus Liberibacter solanacearum (CLso)-infected potato (a) and Candidatus Liberibacter asiaticus (CLas)-infected citrus explants (b). c, d Visual confirmation of GFP expression in hairy roots by fluorescence microscopy. Scale bars, 1 cm. e, f Detection of CLso and CLas in the hairy roots by PCR amplification of diagnostic markers specific to CLso (16S rDNA) and CLas (rplk04/J5 and RNR). GFP, rolB, and rolC encoded on the Ti and Ri plasmids, respectively, and co-transformed into the hairy roots, were used as additional markers for hairy root authenticity; RPL2 and GAPC2 are endogenous potato and citrus genes, respectively, used as genomic DNA controls for PCR. “L” and “HR” indicate leaf and hairy root samples, respectively. “+” indicates positive controls used for the respective PCR amplifications. g, h Temporal growth curves of CLso and CLas in the hairy roots. The Ct values of CLso and CLas in hairy root samples collected at different days in propagation are plotted, whereas the approximate genome equivalents (GE) per nanogram of root genomic DNA are indicated above the bar graph columns. Error bars represent ± standard error of mean (n = 4 and n = 3 for g and h, respectively). Uncropped raw agarose gel images used to prepare e and f are presented in Supplementary Fig. 10. Source data underlying Fig. 1g, h are provided as a Source Data file.