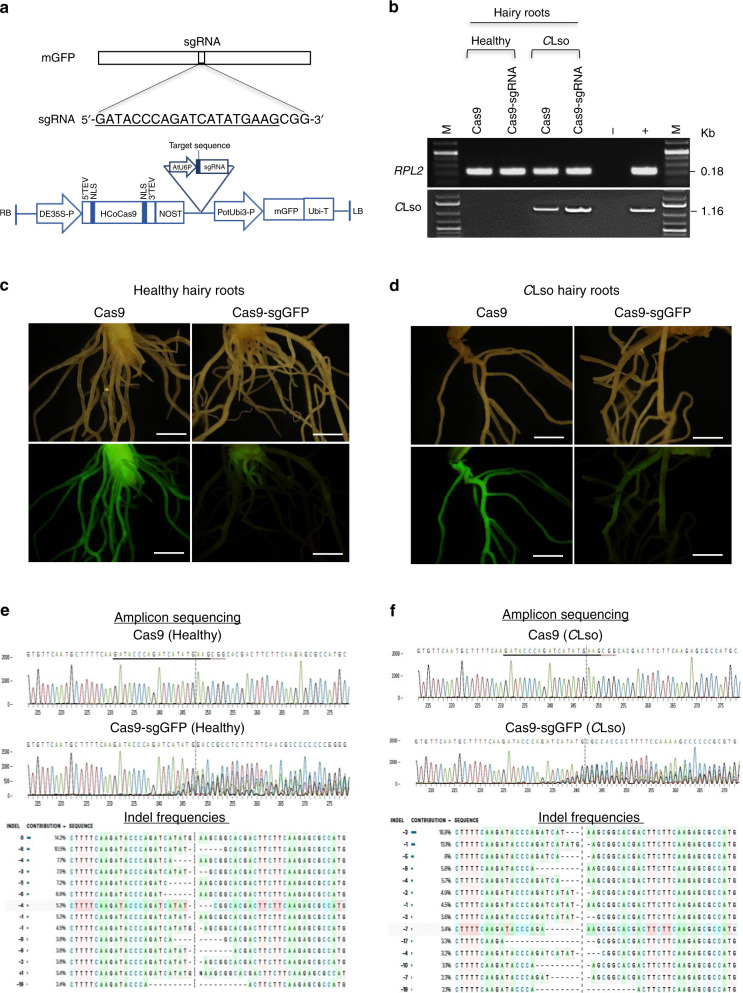
Fig. 3. Evaluation of genome editing in microbial hairy roots.
a Typical CRISPR–Cas9 gene editing construct. The underlined sequence is the target DNA site used in the sgRNA. b–d Transformation of Cas9 alone (control) and Cas9–sgGFP targeting stably expressed GFP transgene in healthy and Candidatus Liberibacter solanacearum (CLso) hairy roots. The loss of GFP fluorescence in Cas9–sgGFP, but not in Cas9 alone, indicates successful editing of the GFP transgene. Scale bars, 1 cm. The experiment was independently repeated two times, and all attempts of replication were successful. e, f Amplicon sequencing confirmed gene editing in the target site (GFP), as indicated by presence of indels in the Cas9–sgGFP hairy roots, but not in Cas9-alone hairy roots. Frequencies of indels detected were ~86 and 100% in the healthy and CLso hairy roots, respectively. Uncropped raw agarose gel images used to prepare Supplementary Fig. 3b are presented in Supplementary Fig. 11.