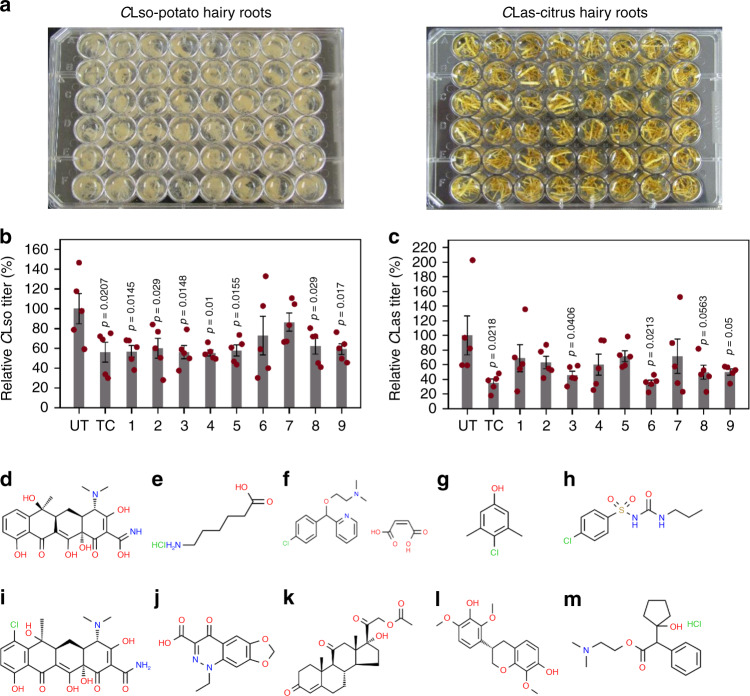
Fig. 4. High-throughput screening and identification of small molecules that confer tolerance to Candidatus Liberibacter spp.
a–c Multi-well microbial hairy root culture plates were used to conduct high-throughput screening of ~220 compounds (Supplementary Dataset 1). The efficacy data for nine selected hit compounds were re-assayed at 10 and 25 µM, respectively, in a, b CLso and a, c CLas hairy roots. Untreated (UT) and tetracycline (TC)-treated hairy roots (at 250 and 500 ppm, respectively, for the CLso and CLas assays) were used as positive and negative controls. The bacterial titers were estimated by qPCR after 72 h of treatment with each compound and plotted relative to those of untreated samples (set to 100%). Error bars represent ± standard error of mean (n = 5). p Values were calculated by two-sample t test (one-tailed) relative to untreated samples. d–m Chemical structures of d tetracycline and the nine hits: e aminocaproic acid (#1), f carbinoxamine maleate (#2), g chloroxylenol (#3), h chlorpropamide (#4), i chlortetracycline (#5), j cinoxacin (#6), k cortisone acetate (#7), l duartin (#8), and m cyclopentolate hydrochloride (#9). The structures of the different chemical compounds were retrieved from the ChemSpider database (http://www.chemspider.com/) (last accessed on 15 October 2020). Source data underlying Fig. 4b, c are provided as a Source Data file.