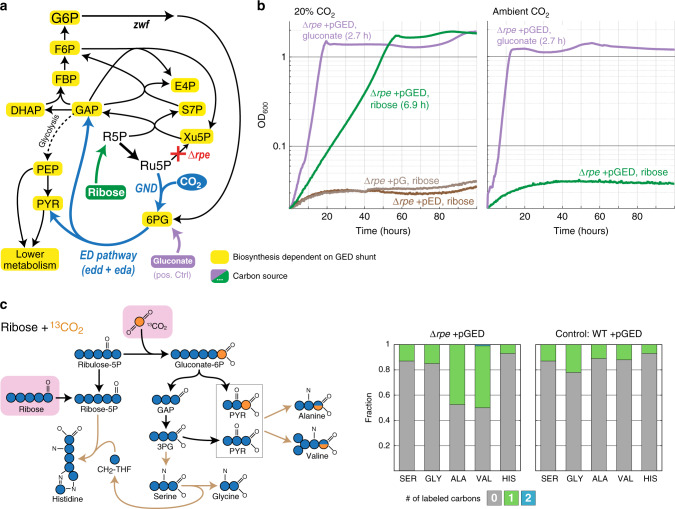
Fig. 2. Activity of the GED shunt in a ∆rpe strain.
a Design of the ∆rpe selection scheme. Ribose can be assimilated only via the activity of the GED shunt, where the biosynthesis of almost all biomass building blocks is dependent on the pathway (marked in yellow). Growth on gluconate (violet) is not dependent on reductive carboxylation via Gnd and thus serves as a positive control. Reaction directionalities are shown as predicted by flux balance analysis. b Growth of a ∆rpe strain on ribose (20 mM) as a sole carbon source is dependent on elevated CO2 concentration (20%, i.e., 200 mbar) and overexpression of gnd, edd, and eda (pGED). Overexpression of only gnd (pG) or only edd and eda (pED) failed to establish growth (less than two doublings). Cultivation at ambient CO2 also failed to achieve growth. Values in parentheses indicate doubling times. Curves represent the average of technical duplicates, which differ from each other by <5%. Growth experiments were repeated independently three times to ensure reproducibility. c Cultivation on 13CO2 confirms the operation of the GED shunt. On the left, a prediction of the labeling pattern of key amino acids is shown. The observed labeling fits the prediction and differs from the WT control cultivated under the same conditions. Labeling of amino acids in the WT strain stems from the natural occurrence of 13C as well as from reactions that exchange cellular carbon with CO2, e.g., the glycine cleavage system and anaplerotic/cataplerotic cycling. Values represent averages of two independent cultures that differ from each other by <10%. 3PG 3-phosphoglycerate, ALA Alanine, GAP glyceraldehyde-3-phosphate, GLY Glycine, HIS Histidine, PYR pyruvate, SER Serine, VAL Valine. Source data underlying b and c are provided as a Source Data file.