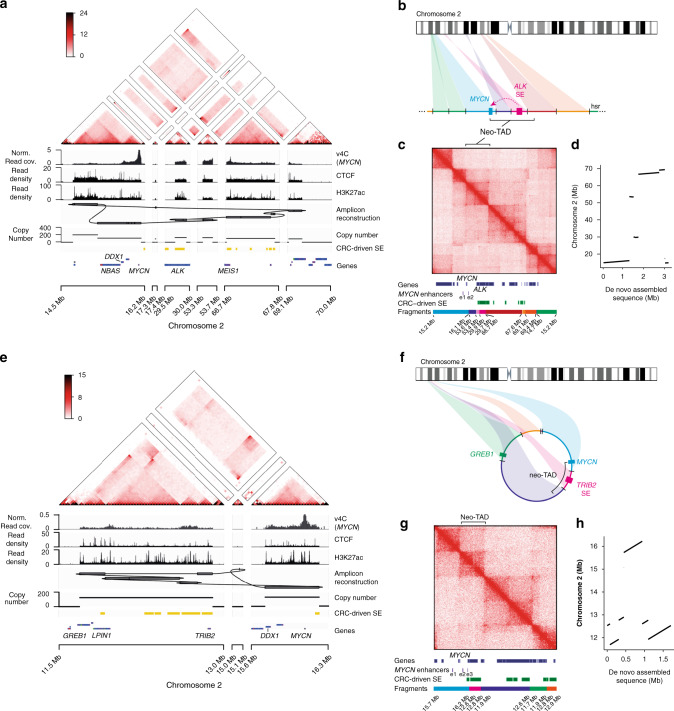
Fig. 4. Reconstruction and epigenetic markup of class II MYCN amplicons.
a, e Short-read-based reconstruction and epigenomic characterization of the MYCN amplicon in IMR-5/75 (a) and CHP-212 (e) cells. Top to bottom: Hi-C map (color indicating Knight–Ruiz normalized read counts in 25 kb bins), virtual 4C (MYCN viewpoint, v4C), CTCF ChIP-seq, H3K27Ac ChIP-seq, Amplicon reconstruction, copy-number profile, super-enhancer locations (yellow), gene positions (blue). b, f Schematic representation of the class II amplicon described in a, e, showing ectopic enhancers and insulator reshuffling leading to locally disrupted regulatory neighborhoods on the HSR in IMR-5/75 (b) and on ecDNA in CHP-212 (f). c, g Alignment of Hi-C reads to the reconstructed MYCN amplicon in IMR-5/75 (c) and CHP-212 (g) and positions of genes, local MYCN enhancers and CRC-driven super-enhancers on the amplicon. d, h Mapping of the long-read sequencing-based de novo assembly of the MYCN amplicon in IMR-5/75 (d) and CHP-212 (h) on chromosome 2. Source data are provided as a Source Data file.