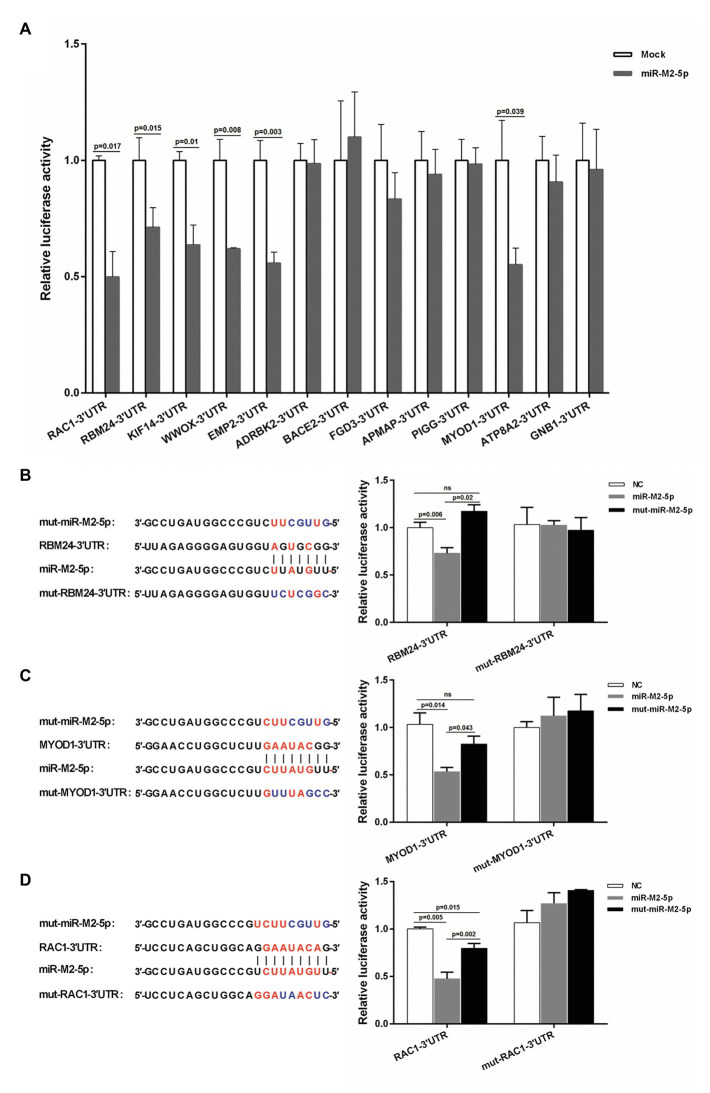
Figure 1.
Interactions between miR-M2-5p and 3'UTRs of candidate mRNA targets determined by dual luciferase reporter assay (DLRA). (A) The first round of DLRA for primary analysis of the interactions between miR-M2-5p and 3'UTRs of 13 mRNA containing perfect base pairing to the seed region of miR-M2-5p, including RBM24, MYOD1, RAC1, ADRBK2, WWOX, EMP2, KIF14, BACE2, FGD3, APMAP, PIGG, ATP8A2, and GNB1. (B–D) DLRA performed for confirming the interactions between miR-M2-5p and 3'UTRs of six distinct mRNA candidate targets, RBM24, MYOD1, and RAC1. The colored seed sequence of miRNA or binding sites of 3'UTRs and the corresponding mutants are shown on the left-hand side. The wild type or mutated 3'UTRs of candidate mRNA targets were cloned into downstream of Renilla luciferase in psiCHECK-2 vector, and then were co-transfected into 293T cells with the miR-M2-5p, mut-miR-M2-5p, or miRNA negative control (NC) mimics, respectively. Firefly and Renilla luciferase activities were measured at 48 h post-transfection (hpt) using the dual luciferase reporter system (Promega). Firefly luciferase was served as the internal control. For each luciferase assay, relative luciferase activity was normalized with respect to miRNA NC. Results are shown as the mean ± SD of three independent experiments. Independent sample t-test was used to analyze the statistical differences between groups. Values of p indicated on columns were used for statistical analyses; ns, no significance.